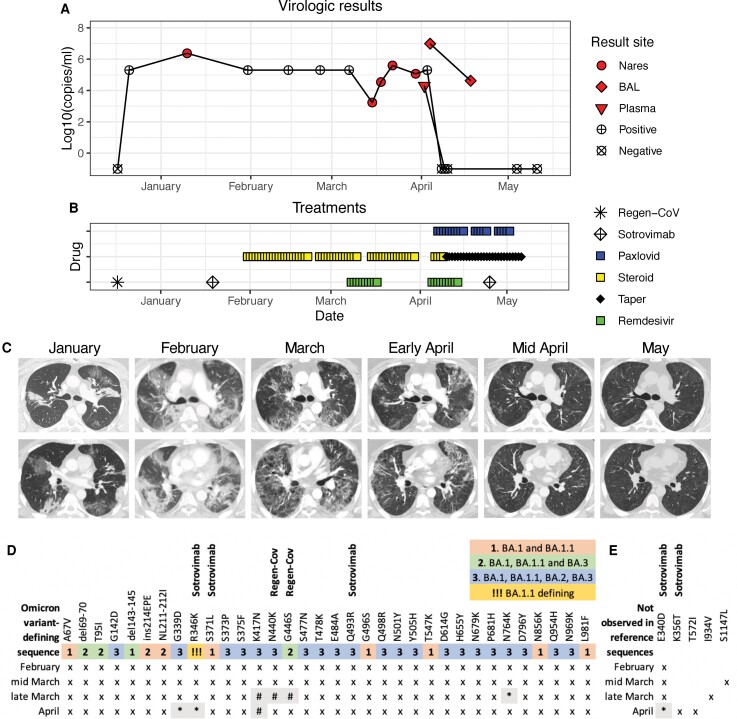
Figure 1.
Virologic test results (A), treatments (B), chest imaging (C), and SARS-CoV-2 spike sequencing (D, E) from exposure to COVID-19 through symptom resolution. A, Filled symbols indicate quantitative results of SARS-CoV-2 PCRs from nasal or nasopharyngeal swabs (circle), bronchoscopy (BAL, diamond) or plasma (triangle). Positive (+) or negative (x) marked symbols indicate qualitative results. Nasal swab was first negative 2 days after starting Paxlovid, and plasma was negative the next day. B, Treatments included three monoclonal infusions (Regen-Cov and Sotrovimab), 2 courses of remdesivir and 20 days of Paxlovid. Steroid doses below 0.3 mg/kg prednisone equivalents are indicated with taper. C, Serial chest imaging by computed tomography (CT). D, Results of SARS-CoV-2 spike sequencing from 3 nares swabs (Feb–March) and BAL (April). Numbers and shading indicate the mutations where each Omicron variant (BA.1-BA.3) differs from the reference sequence NC_045512.2 (Wuhan-Hu-1). Symbols indicate sequencing results at each locus: (x) mutation shown describes the consensus sequence (>50% allelic frequency), (*) mutation describes the consensus sequence in low coverage area (<50 reads), (#) region without sufficient sequencing coverage. Mutations associated with resistance to sotrovimab (E340D, K356T, S371F) and mutations previously seen at high frequency in sotrovimab-treated patients (R346K, Q493R) were observed at multiple timepoints. Mutations associated with escape from REGN10987 (Regen-Cov, N440K, G446S) are present in Omicron variants. E, Non-Omicron-identified consensus mutations. Both E340D and K356T are associated with resistance to sotrovimab. No BA.2-defining sequence changes were observed. Abbreviations: BAL, bronchoalveolar lavage; COVID-19, coronavirus disease 2019; PCR, polymerase chain reaction; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.