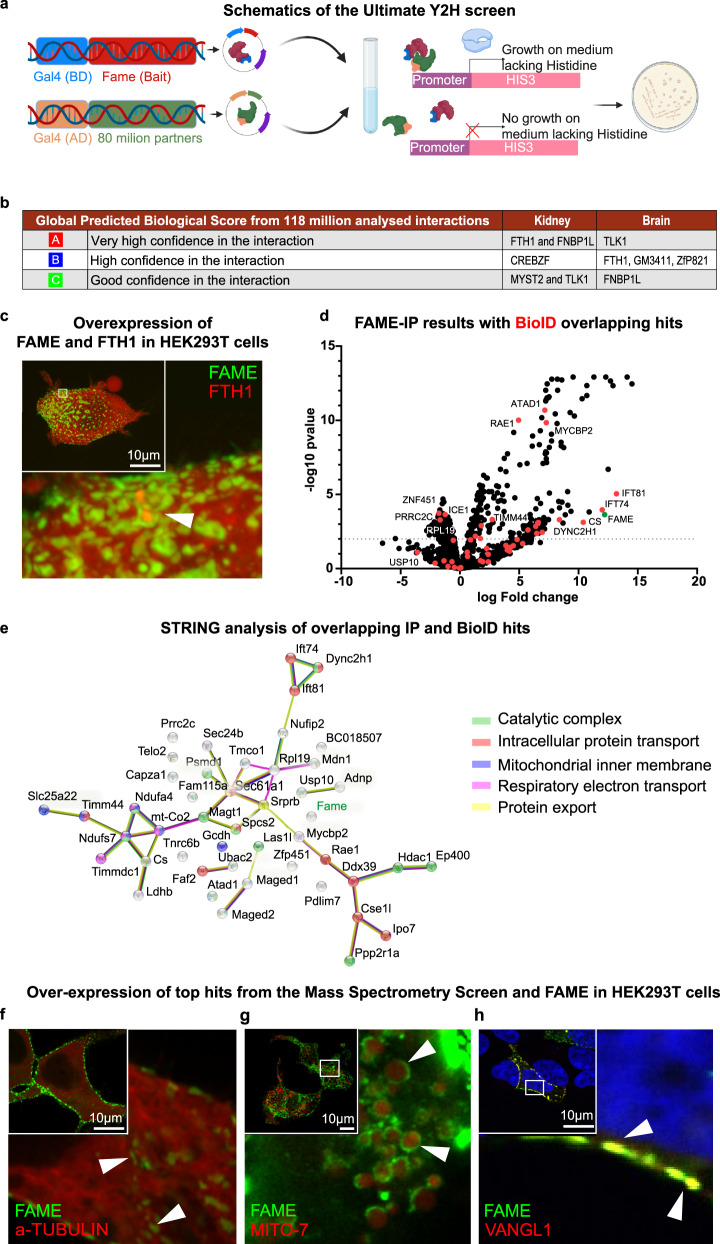
Fig. 3. Binding partners of FAME suggest a role in iron metabolism.
a Graphical representation of the ULTImate Y2H™ screen performed by Hybrigenics. Mouse FAME bait was cloned into the pB27 (N-LexA-AKR2-C fusion) vector, and used for screening using mouse kidney embryo_RP1 and mouse embryo Brain_RP2 fragment libraries as prey. The interaction of two proteins reconstitutes an active transcription factor and enables yeast growth. b Top scoring interaction partners for kidney embryo and embryo brain libraries are indicated. In total, 118 million interactions were tested. c Validation of FTH1 as top FAME interaction partner from the ULTImate Y2H™ screen. FAME-EGFP was overexpressed together with FTH1-mCherry to visualize the heavy subunit of ferritin in HEK293T cells. The white arrow points towards encapsulated FTH1 by FAME. Representative image of 3 independent experiments. d Visualization of FAME interaction partners identified by both immunoprecipitation (IP)/mass spectrometry and proximity-dependent biotin identification analysis (BioID). e STRING analysis from all overlapping IP and BioID hits. f–h Validation of top FAME interaction partners from the mass spectrometry analysis. Representative image of 3 independent experiments. f Overexpression of FAME-EGFP together with α-Tubulin-mCherry in HEK293 cells for the visualization of microtubules. White arrows point at FAME co-localising with α-Tubulin. g Overexpression of FAME-EGFP together with MITO-7-mCherry to identify mitochondria. White arrows highlight the membranous localisation of FAME in mitochondria. h Overexpression of FAME-EGFP and VANGL1-myc visualized with an anti-VANGL1 antibody. White arrows show co-localization of both proteins within the plasma membrane.