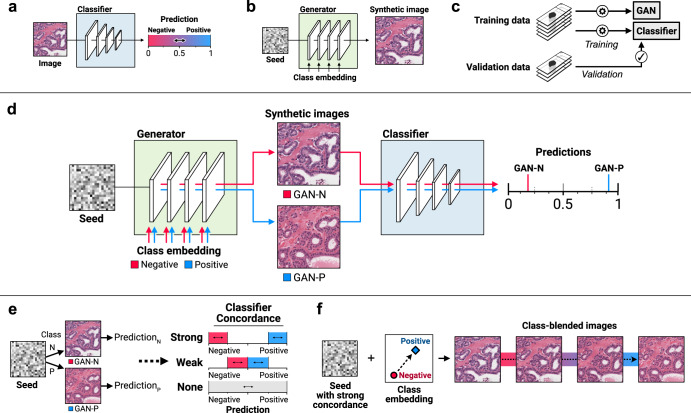
Fig. 1. cGAN-based approach for explaining histopathological models with synthetic histology.
a Our approach starts with a classification model trained to predict a molecular outcome from a histologic image. b A separate conditional generative adversarial network (cGAN) model is then trained to generate synthetic histologic images. cGANs create synthetic images from a seed of random noise and a class label, passed to each layer in the network through an embedding. c The same training data is used for training both the classifier and cGAN. Held-out validation data is used to validate the predictive accuracy of the classifier. d The trained cGAN and classifier are then combined to form a single pipeline. For a given seed, synthetic images are generated for both the negative and positive classes. Both images are then stain normalized and passed to the classifier, resulting in two predictions. If the classifier predictions match the synthetic image labels, the seed is designated “classifier-concordant” and the synthetic images can be used for morphologic explanations. Visualizing classifier-concordant image pairs side-by-side allows one to appreciate the histologic features associated for classifier predictions, providing a tool for model explainability and education. e Classifier-concordant seeds are further subdivided into weakly- and strongly-concordant based on the magnitude of the predictions. f Class-blended images are generated for strongly-concordant seeds by interpolating between class embeddings while holding the seed constant.