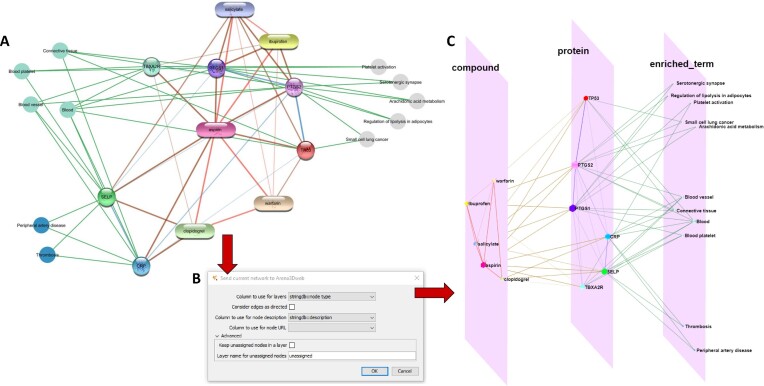
Figure 4.
From Cytoscape to Arena3Dweb. (A) An aspirin chemical-protein interaction network along with enriched terms including diseases, tissues and KEGG pathways generated through stringApp in Cytoscape. Enriched disease nodes are colored red, tissues green and pathways gray. (B) The Arena3DwebApp dialogue window, where users are prompted to choose the node column that will indicate the various layers in Arena3Dweb. Other options include edge directionality, columns for node descriptions and URLs and how to handle nodes without layer information. (C) The generated Arena3Dweb network in three layers; compounds, proteins and enriched terms. Enriched pathways are placed on top, tissues in the middle and diseases on the bottom of the layer. The chemical compound interactions channel is colored red, compound-protein channel in brown, protein-protein in blue and protein participation in the various enriched terms in green. The node coloring scheme is in agreement with the initial STRING network in Cytoscape.