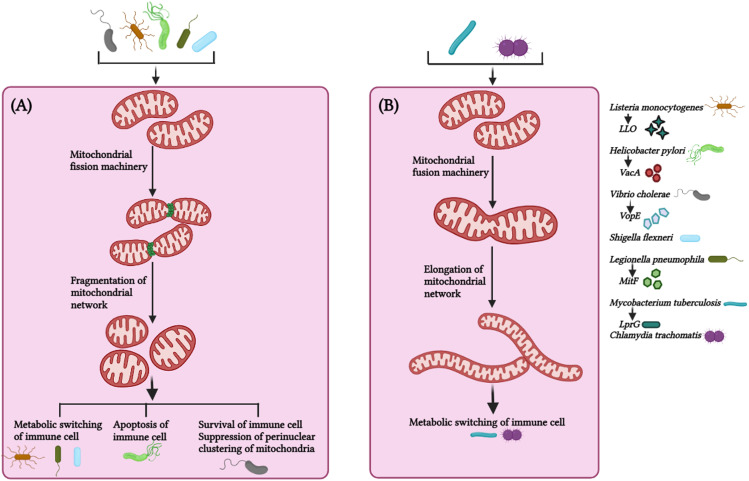
Figure 3.
Mitochondrial dynamics in host immunity against bacterial infection. (A) Bacteria such as Listeria monocytogenes, Helicobacter pylori, Vibro cholerae, Shigella flexneri, Legionella pneumophila triggers fragmentation of the mitochondrial network via virulence factors (LLO – Listeriolysin, VacA – Vacuolating cytotoxin A, VopE – mitochondrial targeting T3SS effector protein, MitF – T4SS effector protein, respectively and effector protein in case of Shigella flexneri is not known) to maintain a favorable microenvironment for pathogen survival and thus promote bacterial survival, persistence and metabolic switching of the immune cells. (B) Bacteria such as Mycobacterium tuberculosis and Chlamydia trachomatis circumvent host immune response by manipulating the mitochondrial dynamics and maintains elongated network of mitochondria at initial phase of infection for their survival. Alteration in mitochondrial fission (by L. monocytogenes, H. pylori, V. cholerae, S. flexneri, L. pneumophila) and mitochondrial fusion (M. tuberculosis, C. trachomatis) suppress bacterial replication and facilitate bacterial clearance from the cell. M. tuberculosis utilize LprG (Lipoarabinomannan carrier protein) for mitochondrial fusion. The image was created with the help of BioRender.com.