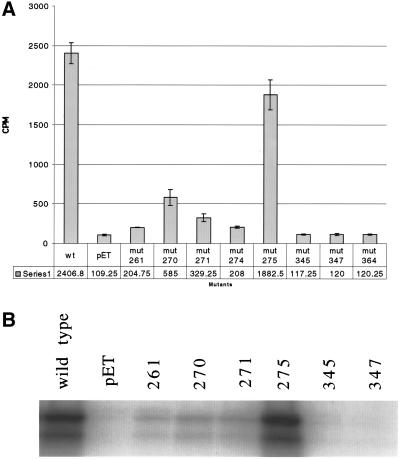
Figure 2.
Analysis of Sir2p's enzymatic activities. (A) Tritiated H4 peptide was used to measure the efficiency of the mutants in an NAD+-dependent deacetylation reaction. The peptide was incubated with 1 mM NAD+ and 2 μg of recombinant protein. The graph measures deacetylation activity by counting the amount of tritiated acetate released from the peptide. (B) Capability of the mutant rSir2 proteins to ribosylate histone H3 was measured by incubating recombinant enzyme with [32P]NAD+ and calf histone H3. The products were run out on a polyacrylamide gel and the gel was exposed to film to see whether the histones incorporated label from the [32P]NAD+.