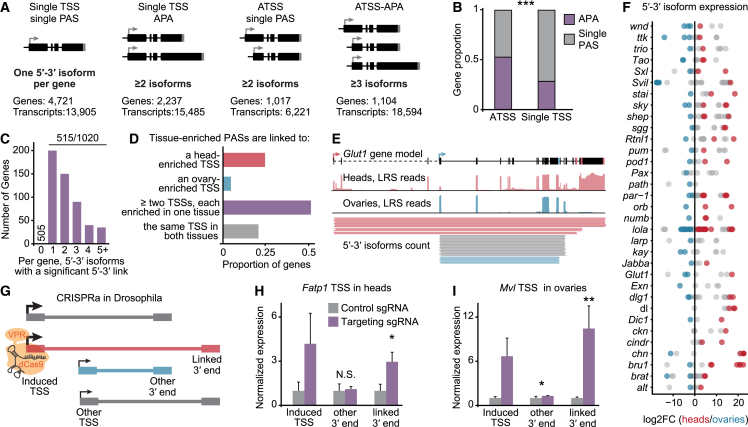
Figure 2.
Transcription start sites drive tissue-specific 3′ end expression
(A) Gene categorization according to TSSs and PASs detected in CIA full-length isoforms. 5′-3′ isoforms were considered distinct if they differed by more than 50 nt at the 5′ end or 150 nt at the 3′ end. The use of several TSSs (arrows) and PASs (stripes) characterizes ATSS and APA genes, respectively.
(B) Proportion of genes that undergo APA in each TSS category. ∗∗∗p < 0.001 (two-tailed Fisher’s exact test).
(C) Number of 5′-3′ isoforms that show a significant difference in expression between heads and ovaries per gene, for all 1,020 ATSS-APA genes expressed in both tissues. Significant links were determined as: |log2FC (5′-3′ isoform expression)| > 1.5 and adj. p value < 0.01 (Wald test, 3 replicates per tissue).
(D) Proportion of genes in which 3′ ends that are enriched in one tissue over the other are associated with a TSS enriched in the same tissue (|log2FC| > 0.5 and adj. p value < 0.05, Wald test, 3 replicates per tissue). Purple indicates that enrichment occurs in both tissues (bidirectional) for at least two PASs.
(E) Glut1 genomic alignment coverage tracks for long reads from heads and ovaries, and depiction of full-length reads representing distinct 5′-3′ isoforms. Read counts per isoform are shown to scale, but each line represents multiple reads. Significant 5′-3′ links are colored in red (heads) and blue (ovaries). Isoforms represented in gray are found in both tissues. Some introns (dashed lines) are not drawn to scale.
(F) Differential isoform expression in Drosophila heads compared with ovaries for a panel of genes that display bidirectional 5′-3′ isoform regulation. Isoforms with a significant 5′-3′ link are colored blue (ovary link) and red (head link).
(G) CRISPRa in fly tissues. Each fly expresses sgRNAs complementary to a tissue-enriched TSS. Association of the sgRNA with a co-expressed dCas9 protein, fused with the transcriptional activator VPR, induces gene activation at the target TSS (red). 5′-3′ isoforms are represented as boxes joined by a straight line; significant links are colored.
(H and I) RT-qPCR quantification of the indicated transcript regions in flies in which dCas9-VPR was recruited to nervous-system TSSs for activation (purple). In control flies, dCas9-VPR was co-expressed with a non-targeting sgRNA (gray). TSS activation of two representative genes, Fatty acid transport protein 1 (Fatp1) and Malvolio (Mvl) is shown in heads (H) and ovaries (I). RNA levels were normalized to RpL32 mRNA, and levels in control flies were set to the value 1. Error bars represent mean ± SD of four biological replicates for each genotype and tissue. ∗p < 0.05 and ∗∗p < 0.01 (one-tailed Student’s t test).