Figure 6.
Dominant promoters drive PAS selection in human brain organoids
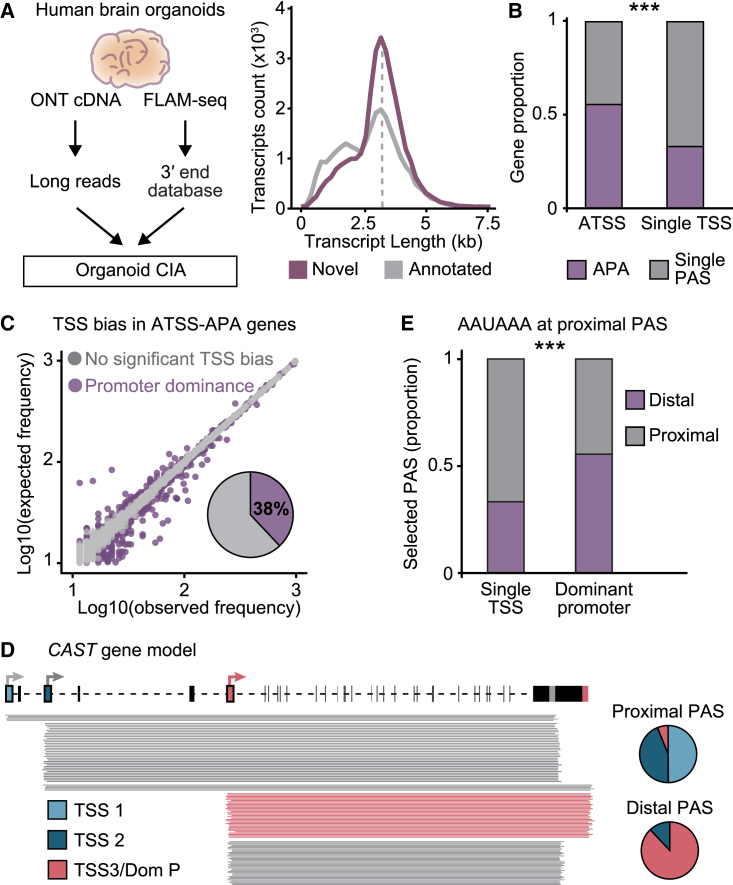
(A) Organoid CIA assembly pipeline. The distribution of novel and previously annotated isoforms as a function of transcript length is indicated (n = 3).
(B) Proportion of genes that undergo APA in each TSS category. ∗∗∗p < 0.001 (two-tailed Fisher’s exact test).
(C) Identification of TSS biases in human brain organoids. The plot was generated as in Figure 3C. 38% of ATSS-APA genes show a significant bias (p < 0.01, chi-squared test with Monte Carlo simulation and Benjamini-Hochberg correction).
(D) Representative example of a gene with promoter dominance in brain organoids. Full-length reads represent distinct 5′-3′ isoforms of the gene CAST. The dominant promoter, its associated PAS, and long reads of the corresponding 5′-3′ isoform are colored in red in the gene model. Pie charts represent the contributions of each TSS to the expression of each 3′ end.
(E) PAS usage when the canonical poly(A) signal AAUAAA is found within a 50-nt window of the most proximal PAS of the gene. Proximal and distal denote PASs found in the proximal 20% or distal 80% of the 3′ UTR, respectively. ∗∗∗p < 0.001 (two-tailed Fisher’s exact test).
see also Figure S6 and Tables S1–S3 and S4.