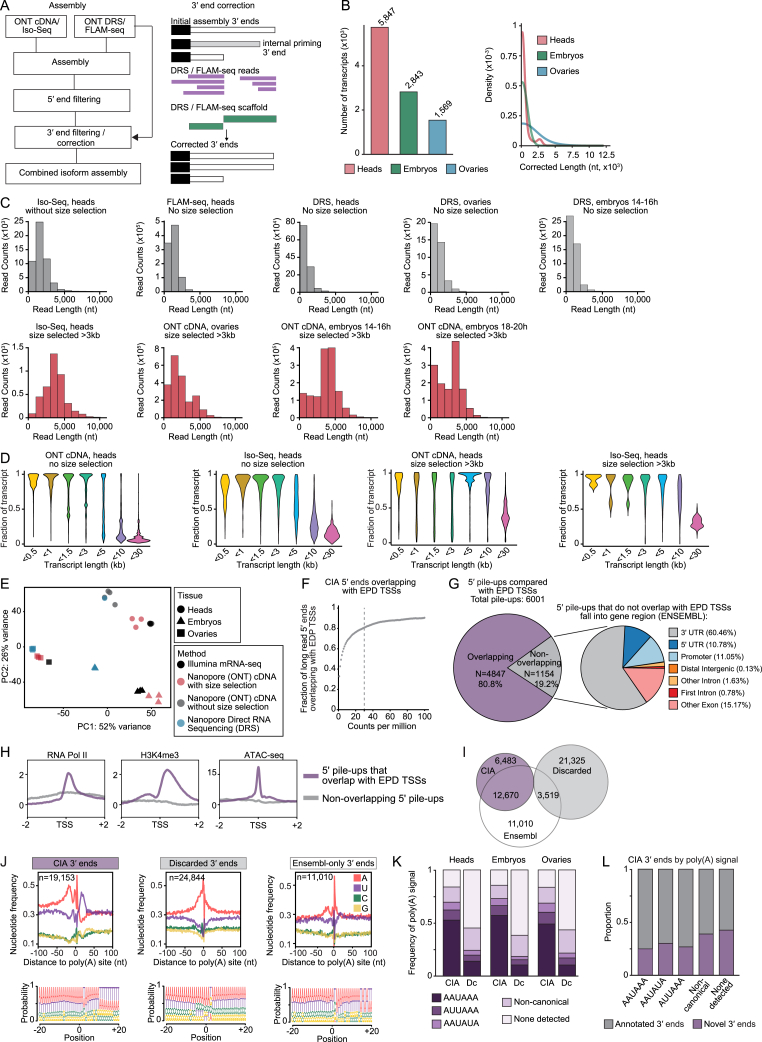
Figure S1.
An accurate, comprehensive, full-length Drosophila transcriptome, related to Figure 1
(A) Combined isoform assembly (CIA) workflow, and schematic of 3′ end correction and filtering. ONT DRS (in heads: ONT DRS and FLAM-seq) data were used to build a database of confident 3′ ends. The CIA assembly was performed with ONT cDNA and ONT DRS (in heads: ONT cDNA, Iso-seq, ONT DRS, and FLAM-seq) data using FLAIR40 and the Eukaryotic Promoter Database (EPD-new).39 Note that due to the low number of Iso-seq reads compared with ONT cDNA reads, Iso-seq reads contribute to CIA to a much lower extent. Since this assembly contains 3′ end artifacts, we filtered out any transcripts with 3′ ends not represented in the DRS/FLAM 3′ end database. Assembled transcript models were corrected with DRS/FLAM 3′ ends.
(B) Number of corrected transcripts per tissue (left) and average length of correction (right). Data from the two embryo datasets (14–16 h AEL and 18–20 h AEL) were pooled.
(C) Read lengths in each tissue with each LRS method. BluePippin size selection (red graphs, below) considerably increased full-length transcript coverage.
(D) Full-length transcript coverage per read for nanopore cDNA and PacBio Iso-seq in heads, before (left two graphs) and after (right two graphs) size selection. For each read, the fraction of the target transcript covered is shown; reads were grouped by the length of the target transcript.
(E) Principal-component analysis plot of gene expression across the samples (3 biological replicates each) generated using nanopore cDNA with and without size selection, nanopore direct RNA sequencing (DRS), and Illumina short-read mRNA-seq from three tissues. Data from the two embryo datasets (14–16 h AEL and 18–20 h AEL) were pooled. Note that LRS methods without size selection (ONT cDNA and DRS, blue and gray) cluster further from mRNA-seq expression estimates (black) than ONT cDNA with size selection (red).
(F) Cumulative plot representing the fraction of long-read 5′ ends that overlap with a TSS described in the Eukaryotic Promoter Database39 in a window of 50 nt, as a function of long-read 5′ end counts per million. A 5′ pile-up was defined as a cluster of >30 counts per million per window (dashed line).
(G) Pie chart representing the number and proportion of 5′ pile-ups that overlap (purple) with a TSS described in the Eukaryotic Promoter Database.39 Non-overlapping pile-ups (gray in the left pie chart) were assessed for the gene region of occurrence (right) as annotated in ENSEMBL.
(H) Cumulative enrichment plots of RNA Pol II ChIP-seq signal, H3K4me3 ChIP signal, and ATAC-seq signal detected at 5′ pile-ups (±2 kb) that overlapped (purple) or not (gray) with TSSs annotated in the Eukaryotic Promoter Database. ChIP-seq and ATAC-seq data from Drosophila heads are from modENCODE.61,49
(I) Venn diagram describing the overlap of mRNA 3′ ends of LRS reads after filtering (CIA) with filtered-out 3′ ends (discarded) and Ensembl-annotated mRNA 3′ ends. 3′ ends detected by FLAM-seq or DRS represent CIA 3′ends (purple); not-detected 3′ ends (gray) were discarded.
(J) Nucleotide composition profiles (spanning 200 nt, top) and sequence logos (spanning 40 nt, bottom) of LRS reads at the cleavage site for each denoted category of 3′ ends from our processing pipeline. Noisy, A-rich distributions are indicative of internal priming. The left and middle panel nucleotide distribution profiles are also shown in Figure 1 and reproduced here for side-by-side comparison with the Ensembl-only category.
(K) In each tissue, proportion of 3′ ends at which the indicated poly(A) signals were detected for each category (CIA or discarded [Dc]). Data from the two embryo datasets (14–16 h AEL and 18–20 h AEL) were pooled.
(L) In CIA transcripts, proportion of 3′ ends carrying a novel (purple) or a previously annotated (gray) 3′ end. CIA transcripts were categorized by poly(A) signal.
Replicates per tissue: ONT cDNA: heads, n = 6; embryos 14–16 h, n = 3; embryos 18–20 h, n = 3; ovaries n = 3. FLAM-seq and Iso-seq: heads, n = 3; DRS: heads, n = 1, embryos 14–16 h, n = 3; ovaries, n = 3. Illumina TrueSeq mRNA-seq: each tissue, n = 2.