Figure S3.
Dominant promoters drive PAS choices, related to Figure 3
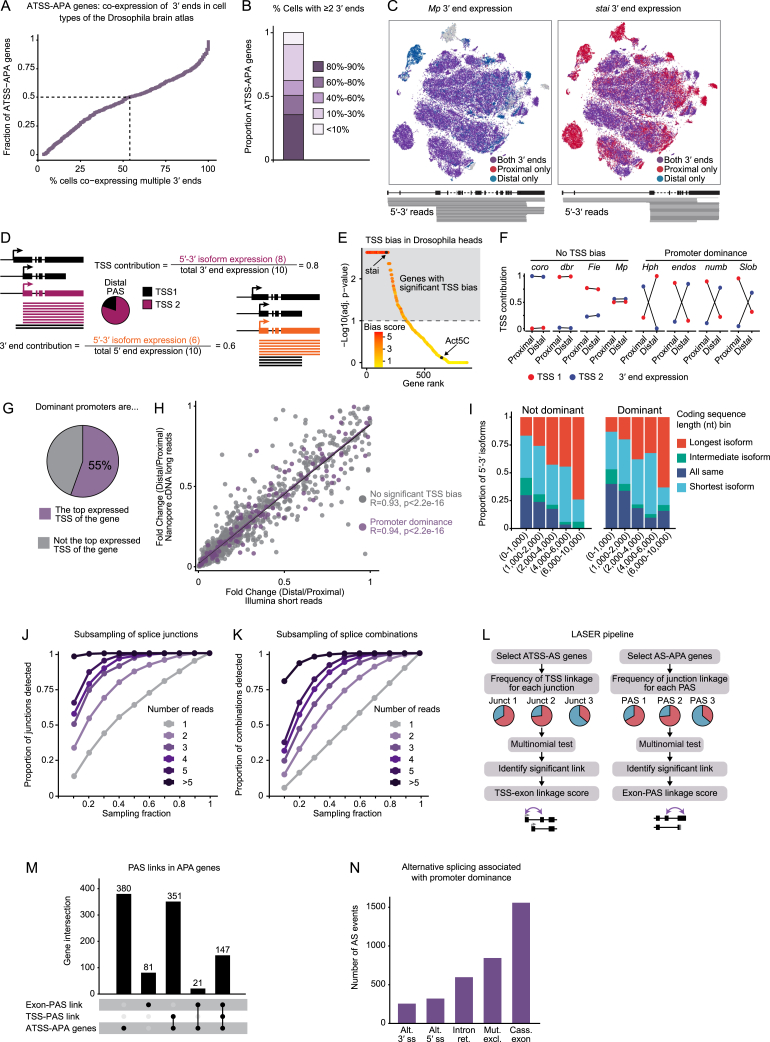
(A) Cumulative plot representing the fraction of ATSS-APA genes as a function of the fraction of cells co-expressing more than one 3′ end across cell types with an expression of more than 0.1 normalized counts.
(B) Proportion of ATSS-APA genes in which two or more isoforms were found to be expressed in the indicated percentage of cells in single-cell RNA-seq data from the Drosophila Brain Atlas.49 For most genes (above the 0.5 proportion), most (over 50%) cells express two or more 3′ ends.
(C) t-distributed stochastic neighbor embedding (t-SNE) maps representing 3′ end expression in Drosophila brain cell types for the two representative genes Multiplexin (Mp) and stathmin (stai). Cells are colored according to their expression of either only the proximal (red), only the distal (blue), or both 3′ end isoforms (purple). Shown below are the gene model and 5′-3′ reads representing the expression of the detected 5′-3′ isoforms. Some introns (dashed lines in the gene model) are not drawn to scale.
(D) Schematic representation of how TSS and 3′ end contributions to 5′-3′ isoform expression were calculated. Full-length 5′-3′ reads were quantified and assigned to 5′-3′ isoforms. For a given 3′ end, the contribution of each 5′-3′ isoform to the expression of the 3′ end was calculated (pink), as well as for a given TSS, the contribution of each 5′-3′ isoform to the expression of the 5′ end (orange). A TSS is termed a dominant promoter for a 3′ end if the respective 5′-3′ isoform expression has a contribution to 3′ end expression significantly higher (p < 0.1, chi-squared test with Monte Carlo simulation and Benjamini-Hochberg correction, also see E) than that of all other 5′-3′ isoforms for the same 3′ end.
(E) TSS bias in ATSS-APA genes assessed using multinomial testing in Drosophila heads. The observed vs. expected counts of 5′-3′ isoforms were used for multinomial testing (chi-squared test with Monte Carlo simulation and Benjamini-Hochberg correction, n = 3). Genes are represented as dots, ranked by p value and color-coded according to bias score (promoter dominance score: absolute value of residuals). Highest-ranked genes (220 genes in the brain) represent near-exclusive 5′-3′ combinations, as exemplified by stai.
(F) Promoter dominance and absence thereof (no TSS bias) shown on representative ATSS-APA genes with two TSSs and two PASs. The proportional contribution of the first TSS (red) and the second TSS (blue) to the expression of the proximal and the distal 3′ end of the same gene are indicated. Lines crossing signify TSS contributions that differ significantly between the PASs.
(G) Pie chart representing the percentage of dominant promoters that constitute the top expressed TSS of the gene in heads.
(H) Scatterplot showing the expression ratio between isoforms expressing the distal and proximal PAS, respectively, measured by long-read sequencing (ONT cDNA), in function of ratios measured by mRNA-seq (Illumina short reads). The ratios were calculated by estimating the ratio of normalized TPM (transcripts per million) assigned to proximal and distal 3′ ends in APA genes. Each dot represents a gene. The correlation coefficient (two-tailed Pearson correlation) is indicated for genes with a dominant promoter (promoter dominance) and TSS-unbiased genes (no significant TSS bias).
(I) Proportion of 5′-3′ isoforms by category, expressing the indicated types of coding sequence, as a function of coding sequence length. Coding sequences are categorized by length within the gene context and represent either the longest, shortest, or an intermediate CDS isoform. Coding sequences of a gene were considered of identical length (all same) if none differed by more than 200 nt. 5′-3′ isoforms are grouped into 5′-3 isoforms with a dominant promoter (dominant) and 5′-3′ isoforms with no dominant promoter (not dominant).
(J and K) Saturation analysis of splice junctions (J) and splice combinations (K) in CIA transcripts, grouped by their expression in number of reads. Reads were randomly sampled in the indicated fractions and a junctions (J) or combinations (K) database was built for each fraction. Splice junctions are exon-exon junctions. Splice combinations are unique assemblies of consecutive exons for each gene. Exons containing, or upstream of, a TSS (first exon), or containing or downstream of a PAS (last exon), were excluded from the analysis.
(L) Long-reads-based alternative splicing estimation and recognition (LASER) framework to identify TSS biases in alternatively spliced (AS) genes (left), and splicing biases in alternatively polyadenylated (APA) genes (right). TSS-exon bias: for each splice junction of each ATSS-AS gene, the observed vs. expected frequencies of TSS-junction combinations were calculated to identify TSSs disproportionately associated with the junction (TSS-exon links). Exon-PAS bias: for each PAS of each AS-APA gene, the observed vs. expected frequencies of splice junction-PAS combination were calculated to identify splice junctions disproportionately associated with the PAS (exon-PAS links). Significant TSS-exon and exon-PAS links were identified by multinomial testing (p < 0.1, chi-squared test with Monte Carlo simulation and Benjamini-Hochberg correction) and assigned a linkage score (sum of squares of residuals). Splice junctions are exon-exon junctions.
(M) Genes in which alternative polyadenylation is linked to alternative splicing (exon-PAS links) or transcription start sites (TSS-PAS link: promoter dominance), or both. Intersections between the gene sets are depicted as connecting lines. The number of genes in each exclusive group is indicated. Only 81 genes with an exon-PAS link were identified outside of the ATSS-APA gene group, and only 21 within the ATSS-APA gene group that were not associated with a dominant promoter (TSS-PAS link).
(N) Type and number of alternative splicing events found in mRNA isoforms transcribed from dominant promoters: alternative 3′ splice site; alternative 5′ splice site; intron retention; mutually exclusive exon; cassette exon.