Abstract
Because of the pervasiveness, persistence, and toxicity of per- and polyfluoroalkyl substances (PFAS), there is growing concern over PFAS contamination, exposures, and health effects. The diversity of potential PFAS is astounding, with nearly 10,000 PFAS catalogued in databases to date (and growing). The ability to detect the thousands of known PFAS, and discover previously uncatalogued PFAS, is necessary to understand the scope of PFAS contamination and to identify appropriate remediation and regulatory solutions. Current non-targeted methods for PFAS analysis require manual curation and are time-consuming, prone to error, and not comprehensive. FluoroMatch Flow 2.0 is the first software to cover all steps of data processing for PFAS discovery in liquid chromatography–high-resolution tandem mass spectrometry samples. These steps include feature detection, feature blank filtering, exact mass matching to catalogued PFAS, mass defect filtering, homologous series detection, retention time pattern analysis, class-based MS/MS screening, fragment screening, and predicted MS/MS from SMILES structures. In addition, a comprehensive confidence level criterion is implemented to help users understand annotation certainty and integrate various layers of evidence to reduce overreporting. Applying the software to aqueous film forming foam analysis, we discovered over one thousand likely PFAS including previously unreported species. Furthermore, we were able to filter out 96% of features which were likely not PFAS. FluoroMatch Flow 2 increased coverage of likely PFAS by over tenfold compared to the previous release. This software will enable researchers to better characterize PFAS in the environment and in biological systems.
Keywords: Mass spectrometry, PFAS, Software, Non-targeted analysis, Aqueous film forming foam, Liquid chromatography
Introduction
The diversity of per- and polyfluoroalkyl substances (PFAS) is astounding, making the comprehensive detection of these often trace components of environmental, industrial, and biological matrices challenging. The vast structural diversity of PFAS is matched by their broad range of applications; PFAS are often important components in non-stick cookware, paper and paper board coatings (e.g., food wrappers), firefighting foams, stain repellent sprays and coatings for textiles, carpeting and furniture, aviation hydraulic fluid, mechanical components, surfactants used in mining, paints, inks, adhesives, and biocides, to name a few [1–3]. Their widespread use and often superior performance across many sectors are due to their unique intrinsic properties. For example, the carbon-fluorine (CF) bond is the strongest carbon bond in nature, leading to high durability and the carbon-fluorine chains are both oil and water resistant. These unique properties, along with their ubiquitous application, make PFAS a serious concern from a health perspective. Furthermore, PFAS are highly persistent and are capable of traveling far distances from points of release, and in many cases can end up in drinking water intakes [4, 5]. Many PFAS can bioaccumulate [6–11] and there is a growing list of associations with numerous disease states and metabolic effects [12, 13] including high cholesterol [14, 15] and triglyceride levels [16, 17], thyroid disease [15], pregnancy-induced hypertension [18, 19], ulcerative colitis [20], kidney cancer [21, 22], testicular cancer [21], reduced vaccine response [23], and immunotoxicity [12, 23–25].
Current epidemiological and environmental studies generally target the few well-known PFAS. For example, most large-scale epidemiology studies focus on only perfluorooctanoic acid (PFOA) and perfluorooctanesulfonic acid (PFOS) [12, 14–22, 24, 26]. This is problematic, as infrequently measured or unknown PFAS with yet to be determined toxicities are often present in PFAS mixtures, with possible synergistic or additive effects [27]. In addition, the most commonly screened PFAS may not be representative of the most abundant exposures; in one study, dependent on participant location, 30–70% of the total carbon-fluorine content detected in serum was not attributable to commonly measured PFAS [26]. As of November 2020, the United States Environmental Protection Agency (EPA) has compiled a list of over 8000 PFAS structures [28] made available via their CompTox Chemicals Dashboard [29]. This list is not exhaustive and previous studies have found compounds that are not contained on the list [30]. At the time of writing, the dashboard has less than one million chemicals (~883,000) and other databases such as CAS SciFinder, ChemSpider, and PubChem each contain tens of millions of substances with additional tens of thousands of PFAS. The explicit structural space for PFAS, and what is actually in the environment, especially taking into consideration potential degradation products, is enormous in scope.
The diversity of PFAS in the environment is expected to increase as new PFAS are synthesized, used, and released into the environment and, in some cases, when existing PFAS undergo environmental and biological transformations. Therefore, to determine the potential health effects and scope of PFAS burden, it is important to empower the scientific community with tools capable of the comprehensive detection of these compounds. Non-targeted liquid chromatography–high-resolution tandem mass spectrometry (LC-HRMS/MS) is one tool which can be used to detect a large portion of these chemical species in a single measurement, while also providing important information about the specific compounds present [31]. The current major bottleneck in non-targeted mass spectrometric analysis is in data processing and interpretation. While numerous data processing workflows have been introduced to improve the coverage and accuracy of PFAS annotations [32], use of these LC-HRMS/MS data processing methods is tedious and prone to human error. Furthermore, it is impossible to manually survey the gigabytes of available data to comprehensively and confidently identify the PFAS covered by LC-HRMS/MS. Therefore, we introduce FluoroMatch 2.0, which is the first software to automate the non-targeted identification workflow for PFAS, aiding researchers in compound annotation.
Whereas the original release of FluoroMatch [30] used a suspect screening approach that targeted compounds based on available MS/MS spectra acquired from standards and information available in literature, FluoroMatch 2.0 uses truly nontargeted methods which can be used to enhance the identification and discovery of known, uncommon, and unknown PFAS. FluoroMatch 2.0 introduces an automated workflow which includes blank filtering, mass defect filtering, Kendrick mass defect plots (normalized to CF2) [33], retention time pattern analysis, and exact mass matching to take full advantage of retention time and exact mass data. Furthermore, FluoroMatch 2.0 screens common PFAS fragments and incorporates MS/MS libraries predicted for all PFAS listed in the EPA database that are likely to form [M-H]− ions. This is in addition to the confident MS/MS rule-based annotation [34] derived from PFAS standards in the original FluoroMatch release [30]. All evidence is incorporated into a clear, yet thorough, scoring framework for ease of interpretation, which can be applied across multiple workflows by analytical chemists measuring PFAS. This can harmonize reporting across different laboratories.
Aqueous film forming foams (AFFFs), commonly used to fight fuel fires, are complex mixtures containing hundreds of both well-characterized and novel PFAS [35–37] and thus present an ideal medium for demonstrating the application of FluoroMatch 2.0 to PFAS identification and discovery. Use and storage of AFFFs at military, industrial, and municipal sites have led to extensive ground and drinking water contamination [4, 5, 38–41]. Legacy “Lightwater” electrochemically fluorinated AFFFs were phased out as of 2002 [42], but they remain environmentally relevant, understudied, and a major contributor to ongoing PFAS contamination [41, 43].
Materials and methods
Automated compiling of evidence for PFAS annotation using retention time, exact mass, and fragmentation information
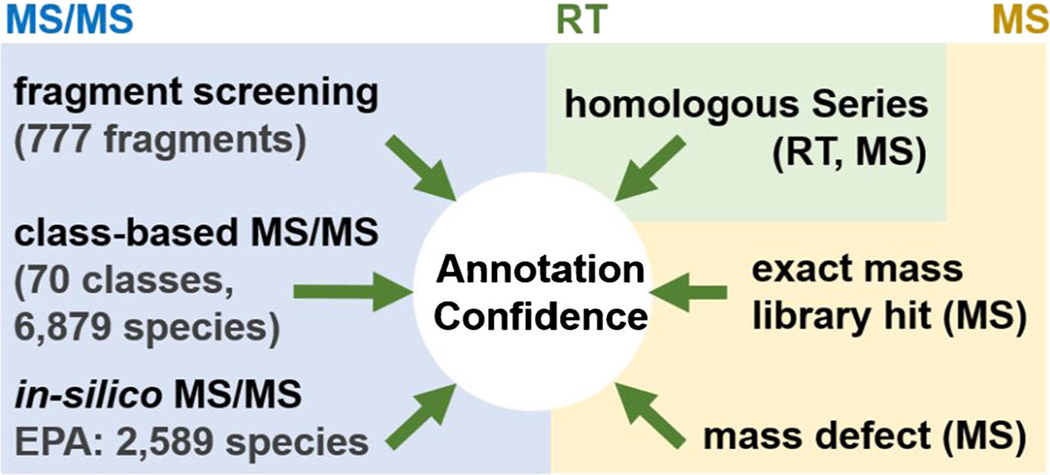
Confidence in annotations is dependent upon the number of orthogonal chemical measurements and how unique they are to the compound of interest. Therefore, in FluoroMatch 2.0, we aimed to include as much evidence as possible in determining PFAS annotations; we used the three most employed characteristics measured in LC-HRMS/MS: exact mass, retention time, and fragmentation (MS/MS) (Fig. 1). FluoroMatch 2.0 determines exact mass matches by searching against the EPA PFAS structure list, simulated structures from standards, and structures reported in literature. The default FluoroMatch 2.0 library includes 9468 molecules that form [M-H]− ions, but any exact mass library can be substituted. Exact mass is further used to flag features which have mass defect values expected for PFAS: those falling between − 0.11 and 0.12. These values are based on the 5th and 95th percentile of the EPA PFAS structure list, meaning that 90% of the features in the list have a mass defect in this range. Note that PFAS containing mercury, iodine, a significant proportion of hydrogens and other elements may not meet this criterion, but may still be annotated using further evidence described below.
Fig. 1.
Evidence used for annotation in FluoroMatch 2.0 (retention time (RT), exact mass from full-scan data (MS), and fragmentation (MS/MS))
Further evidence is gleaned from exact mass by automatic identification of homologous series and their members. All members of a series have the same CF2-normalized mass defect within 0.005 Da (user modifiable) and are separated by a normalized mass difference divisible by 50 Da (CF2 unit separation). Screening across other repeating units (e.g., OCF2) has been implemented as well. Retention time information is used to verify whether compounds fall within a homologous series; a compound is flagged if increasing CF2 units (or other types of repeating units) do not have increasing retention time. Compounds which are flagged are not included when determining the total count of a homologous series used for scoring (Fig. 1).
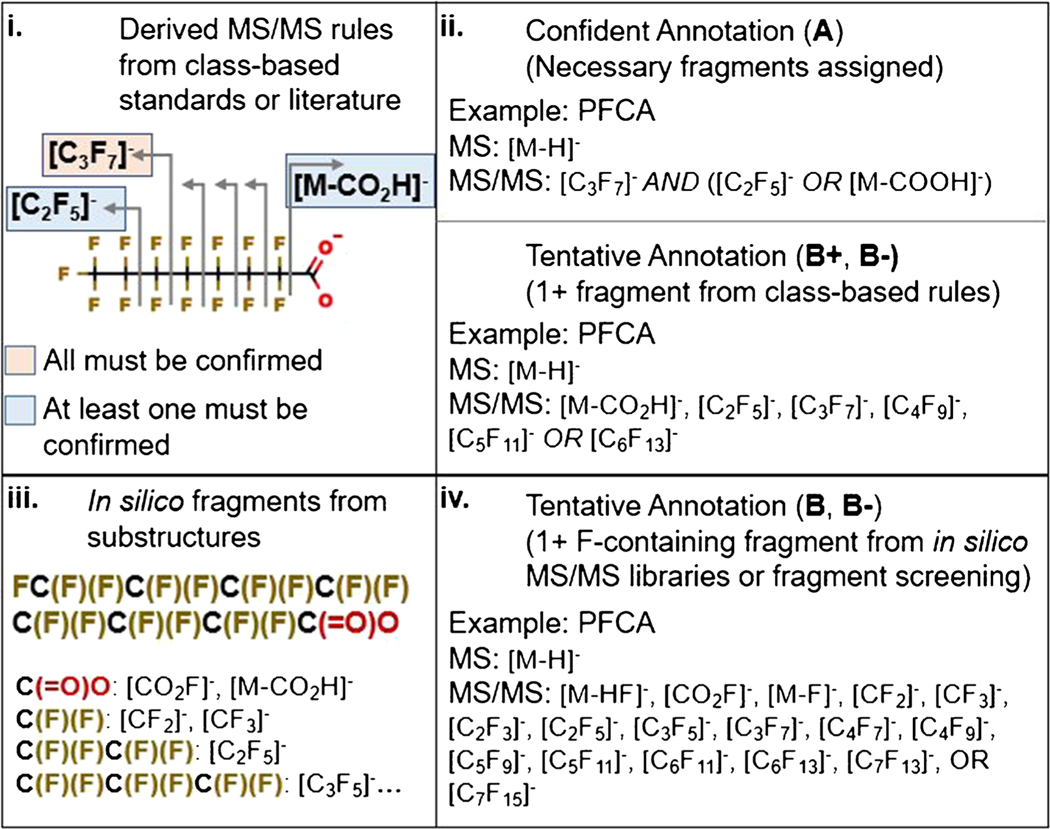
For confident annotations in non-targeted analysis, it is paramount to have fragmentation evidence. In the previous release of FluoroMatch, class-based fragmentation rules were generated (Fig. 2i) which provided highly confident results (Fig. 3), but were limited to compounds for which standards were run within the same PFAS class. We expanded MS/MS coverage to all compounds within the EPA PFAS structure list. We assigned fragment masses to substructures of PFAS molecules using the SMILES (computer and human readable notation for describing chemical structure) in the EPA list. For example, [C2F5]−, [CF3]−, and [CF2]− were assigned to any compound containing the following SMILES substring: C(F)(F)C(F)(F) (Fig. 2iii). Fragment-substructure relationships were determined manually based on careful annotation of MS/MS spectra from PFAS standards and literature and determination of common fragmentation schemes. We developed a library of 728 PFAS substructures with one or more fragments assigned; there are 878 unique fragments in the library to date (777 fluorine containing fragments). The code used to assign fragments, given any list of SMILES structures (SMILES2MSMS.r), is provided with FluoroMatch 2.0.
Fig. 2.
Two different methods for generating MS/MS libraries of PFAS (i and iii) and three different methods for MS/MS annotation are shown (ii and iv). PFAS MS/MS rules are assigned for each PFAS class with standards acquired or annotated spectra from the literature (i), which are used for confident annotations (ii). Furthermore, tentative confident annotations with one or more of these class-based fragments observed are also assigned (ii). “In silico” fragmentation libraries which are more comprehensive in terms of fragment coverage but potentially less accurate are developed by assigning substructure-fragment relationships (iii). In this case, fragmentation is predicted from molecules in the EPA structure list. Tentative annotations using this library are made if F-containing fragments are observed (iv)
Fig. 3.
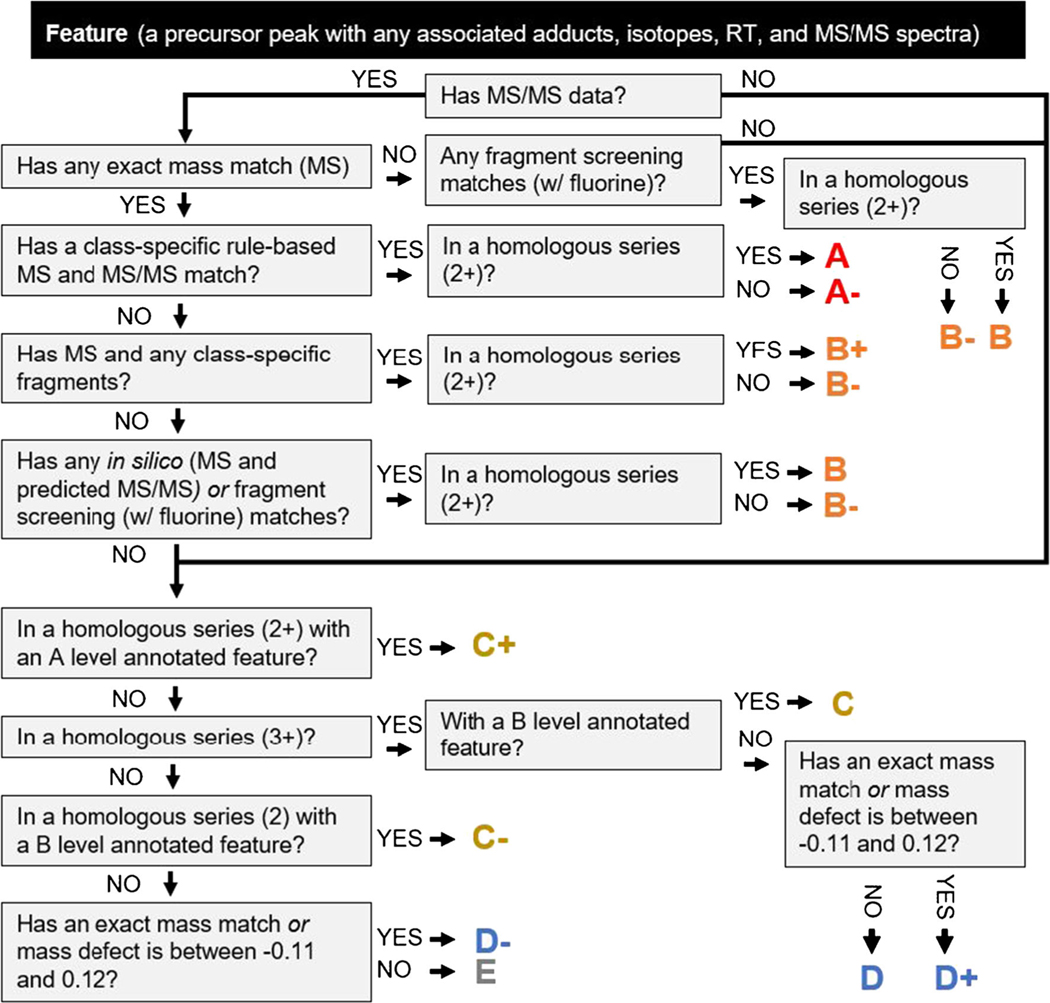
Scoring system implemented in FluoroMatch 2.0. Acronyms: MS—in this case refers to exact mass matches; MS/MS—refers to any algorithms employed by FluoroMatch 2.0 using exact mass to match experimental fragmentation to known PFAS fragments; RT—retention time; homologous series—in this case PFAS in the same class but with a different number of CF2 repeating units determined by exact mass intervals (50 for CF2) and the same CF2-normalized mass defect
Currently, fragment-substructure relationships are assigned (Fig. 3) for only [M-H]− adducts. FluoroMatch 2.0 was built to work with both positive and negative polarity for various adducts, and future work will be done to implement new libraries expanding FluoroMatch coverage. Since fragment masses observed often depend on the adduct selected for fragmentation, we applied the MZedDB [44] to only include SMILES from the structure list which can possibly generate [M-H]− adducts. This reduced the suspect list of PFAS from the EPA list from over 8000 compounds to 2589 unique PFAS species for which fragment masses were predicted for [M-H]− adducts. In this manuscript, these fragment libraries are referred to as in silico MS/MS (Fig. 2iii) as opposed to the fragmentation rules that were developed using class-based MS/MS from standards (Fig. 2i).
Users can generate additional in silico libraries using the SMILES2MSMS.r code contained in the FluoroMatch 2.0 distribution, which only requires a list of SMILES for which the user wants to predict fragmentation. Substructures and their associated fragments are matched to the remaining SMILES of the molecules using the “match” function from the RCDK package [45] in R. Output MS/MS libraries can be directly integrated into the FluoroMatch 2.0 software. Further details on the SMILE2MSMS algorithm can be found in the Supplementary Information (ESM).
In addition to traditional screening, where fragment masses are used to confirm a precursor exact mass match and structural annotation, FluoroMatch 2.0 also incorporates reverse fragment screening (Fig. 1). Any precursor mass falling within a mass defect between − 0.116 and 0.268 (98% of compounds in the EPA structure list) and a mass range of m/z 117 to 1189 (98% of compounds in the EPA structure list) has experimental MS/MS data searched against 777 PFAS fluorine-containing fragments annotated from standards and the literature (note that 878 above includes fragments without fluorine). Based on fragment mass matches, PFAS substructures are assigned, and the precursor ions are flagged as potential PFAS (Fig. 3). The result is a list of potential PFAS features with fragment annotations, though the identities of the precursors may be unknown and have no exact mass match.
Classification of confidence for FluoroMatch 2.0 results: universal schema for harmonization of reporting PFAS annotation evidence
After all pieces of evidence are provided for a feature, it can be difficult to interpret the PFAS annotations in terms of confidence. Therefore, we introduce a detailed scoring system combining all pieces of evidence provided by FluoroMatch 2.0 (Automated compiling of evidence for PFAS annotation using retention time, exact mass, and fragmentation information section). The scoring is broken into 5 general categories (A, B, C, D, and E), with three subcategories each (e.g., B+, B, and B−) (Fig. 3). Additionally, we have provided notes on how our confidence rankings relate to the levels proposed by Schymanski et al., although comparison is challenging. This scoring system is unique compared to the Schymanski et al.’s approach [46], due to specific evidence available for PFAS non-target data (homologous series, mass defect filtering, etc.). While there are a number of subcategories, which makes interpretation and adoption more complex, these subcategories do provide valuable distinctions for interpreting the data. Interpretation is simplified by providing all explanations of assignment confidence in adjacent cells in the output table. Furthermore, interpretation is actionable for the major categories: structural annotations assigned as A can be interpreted as confident without the need for thorough manual validation. In contrast, manual validation is needed for any structural annotations assigned as B for which MS/MS evidence exists but the structure is tentative. Any feature assigned as C is a member of a series which contains MS/MS evidence but the feature itself has no MS/MS evidence; manual validation is thus necessary, but other members of the series can be used to propose a starting structure. In contrast, features assigned a D are possibly a PFAS based on exact mass evidence only (e.g., exact mass match, mass defect filtering, and/or belonging to a PFAS homologous series) without any evidence providing a starting point for structural annotation. The scoring system is outlined in-depth below and in Fig. 3.
Operational definitions of terms used in categories of annotation confidence
Exact mass match—
match to a compound in the EPA PFAS master list or from compounds found as standards or in literature. Includes compounds within homologous series of those previously described, even if the exact species has not been characterized.
Class-specific fragmentation pattern—
In the case of FluoroMatch 2.0, this refers to the topmost abundant fragment m/z values based on annotated MS/MS from standards or literature for the given PFAS class. Intensity information is not required.
Homologous series—
based on Kendrick mass defect (KMD) plots, with masses normalized to CF2, these are series of 2 or more molecules separated by the mass of one or more CF2 units that have the same CF2 normalized mass defect. Homologous series must follow the correct retention time order: molecules with higher molecular weights should elute later assuming reverse phase chromatography. This flag can be toggled off if reverse phase chromatography is not used. Currently, retention time prediction for PFAS is not included, but future work is being performed in this direction to add a tolerance in addition to order.
Categories of annotation confidence
- Confident annotation with a proposed PFAS structure: has an exact mass match and a class-specific fragmentation pattern matched with experimental MS/MS, and/or is confirmed using standards (e.g., targeted approach). May not differentiate between isomeric species with similar fragmentation (e.g., branched versus straight chain).
- A+ = Annotation using in-house standards (not applicable to FluoroMatch 2.0 software, applicable to targeted methods) (Level 1 Schymanski)
- A = Class-specific dominant fragments observed, exact mass match, and 2+ in homologous series (Level 2 Schymanski)
- A− = Class-specific dominant fragments observed and exact mass match; no series (Level 2 Schymanski);
- A highlylikely PFAS often with a possible structure: based on presence of fluorine-containing fragments common to PFAS. A structure may be proposed based on an exact mass match or structure-fragment relationships. Additional information or manual review is necessary to prove the structure (Level 3/5 Schymanski)
- B+ = 1+ PFAS fragments from class-specific rules, exact mass match, and 2+ in homologous series (Level 3 Schymanski)
- B = 1+ common PFAS F-containing fragments observed (in silico prediction or fragment screening), exact mass match or mass defect filtering (− 0.116 and 0.268 (98% of compounds in the EPA structure list)), and 2+ in homologous series (Level 3 or 5 Schymanski)
- B− = 1+ fluorine-containing fragments and exact mass match or mass defect filtering; no series (Level 3 or 5 Schymanski)
- Possible annotation with a proposed PFAS class based on annotation/evidence of other PFAS (level A or B) within the same homologous series. These compounds might be PFAS, but many other non-PFAS with the same mass likely exist and these cannot be ruled out (Level 5 Schymanski)
- C+ = 2+ in homologous series and at least one confident PFAS annotation within homologous series (A or higher grade)
- C = 3+ in homologous series and at least one highly likely PFAS annotation within homologous series (B or higher grade)
- C− = 2+ in homologous series and at least one highly likely PFAS annotation within homologous series (B or higher grade)
- No annotation but possible PFAS based on the presence in a homologous series, exact mass matches, or mass defect filtering. These compounds might be PFAS but structural information is not known due to no MS/MS information for any members of the series. More information is needed for identification (Level 5 Schymanski)
- D+ = Mass defect between − 0.11 and 0.12 OR exact mass match, and 3+ within homologous series
- D = 3+ within homologous series
- D− = Mass defect between − 0.11 and 0.12 OR exact mass match; no series
Likely not a PFAS: No evidence of PFAS fragments, exact mass matches, homologues series, or a mass defect within the range of most PFAS (Level 5 Schymanski).
Sample preparation, data acquisition, and data processing
An aliquot of a mixed AFFF product collected from a holding tank at a field site in 1999 was prepared by 100,000× dilution in 70:30 water:methanol (Fisher Scientific Optima® LCMS-grade). Blanks consisting of 70:30 water:methanol were run before and after AFFF injections. The AFFF sample was characterized previously and is known to contain primarily electrochemically fluorinated 3M Light Water AFFF mixed with some fluorotelomer-based AFFF [11]. This AFFF sample is representative of mixtures that are widespread at historical AFFF-contaminated sites [43]. The diluted sample was injected four times for iterative exclusion information-dependent analysis (Iterative MS/MS) at 50 μL onto an Agilent 1290 Infinity II ultra-high-performance liquid chromatography (UHPLC) system connected to an Agilent 6545 quadrupole time-of-flight mass spectrometer (Q-TOF MS). The chromatographic method was adapted from previous work [11]. PFAS were detected in negative electrospray ionization mode. Source parameters are listed in ESM Table S1.
All data processing steps were performed using FluoroMatch Flow 2.0. As in the original FluoroMatch release [30], FluoroMatch Flow is the automated workflow referred to throughout, whereas FluoroMatch Modular is only for annotation and scoring and can be integrated with a user’s peak picking workflow. Blank filtering was performed using three solvent blanks and a threshold of the maximum sample being higher than the average of the blanks plus three-times the standard deviation of the blanks. A precursor mass tolerance of ± 0.005 Da was used and a MS/MS mass accuracy of ± 7.5 ppm. A minimum intensity of 60 was used for the MS/MS intensity threshold. All other parameters remained at default values.
Results and discussion
FluoroMatch 2.0 user workflow, processing steps, and libraries
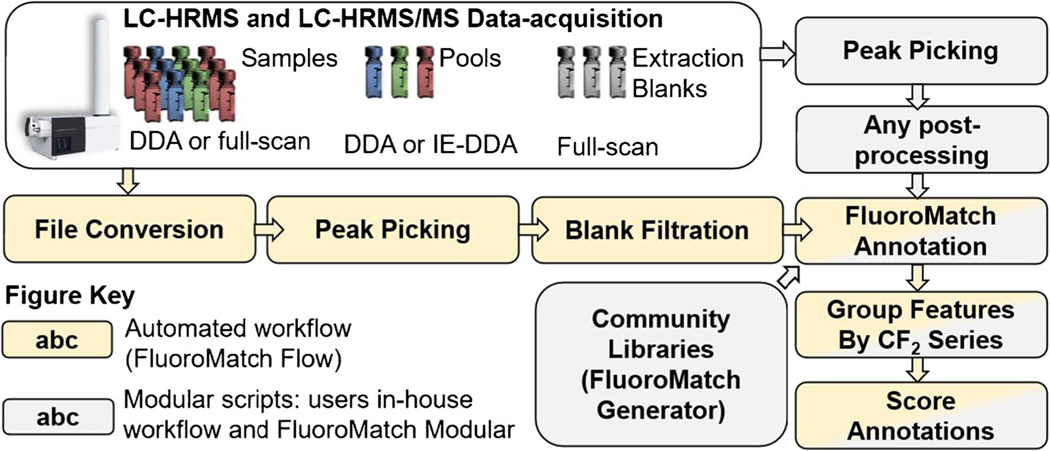
FluoroMatch 2.0 is the first software to automate a PFAS non-targeted analysis workflow. The workflow is user-friendly: for FluoroMatch Flow, users simply drag vendor files onto the interface. The following files are required: at least one full-scan data file (can be data-dependent analysis or other scan type as long as full-scan was acquired and enough scans are found across the peak) and one data file containing quadrupole-selected MS/MS (e.g., data-dependent analysis, iterative exclusion data-dependent analysis, or targeted MS/MS). Ideally, at least three experimental/extraction blanks are included as well (Fig. 4). A modular version of FluoroMatch is also included in the FluoroMatch 2.0 release; this version can be used for annotating feature tables derived using other peak picking and post-processing software (Fig. 4). In-depth tutorials for users are provided in the release, including both a written manual and video tutorials available at: http://innovativeomics.com/software/fluoromatch-flow-covers-entire-pfas-workflow/.
Fig. 4.
FluoroMatch 2.0 user workflow and data processing steps covered by the fully automated version (FluoroMatch Flow) and the modular version (FluoroMatch Modular). Acronyms: DDA, data-dependent analysis; IE, iterative exclusion; LC-HRMS, liquid chromatography–high-resolution mass spectrometry; MS/MS, tandem mass spectrometry. Tan and gray boxes indicate aspects of both automated and modular workflows
To screen PFAS, we compiled and developed one of the largest PFAS public databases to date (Fig. 1), which contains SMILES structures, exact masses, formulae, and predicted or derived MS/MS masses (fragment exact mass only, no intensity) using PFAS-specific fragment-substructure relationships. The database contains 9468 PFAS and non-PFAS fluorinated structures which form [M-H]− adducts. This database includes 2589 unique PFAS and non-PFAS fluorinated species from the EPA structure list which are predicted to form [M-H]− adducts and contains predicted MS/MS using fragment-structure relationships for each of these species. Furthermore, 6879 PFAS structures and their MS/MS were derived from 70 homologous series reported in the literature or for which standards were available, noting that if one or more species had annotated spectra, all species in that series were simulated up to a maximum of 20 CF2 repeating units. In total, 777 unique fluorine-containing fragments were derived from spectra or assumed based on fragmentation patterns observed across different repeating CF2 units. These fragments were used for screening to determine likely PFAS and other fluorinated species and help in annotation. It is important to note that the fragment screening approach will pull out many fragments which may relate to fluorinated compounds only containing one or a few fluorine’s which may be non-PFAS like, for example, [M-F]−, [M-H3F2]−, [SO3F]−, [SO2F]−, and [CO2F]−, and hence the software is not limited to only PFAS species.
Data reduction and confidence assignments using FluoroMatch 2.0
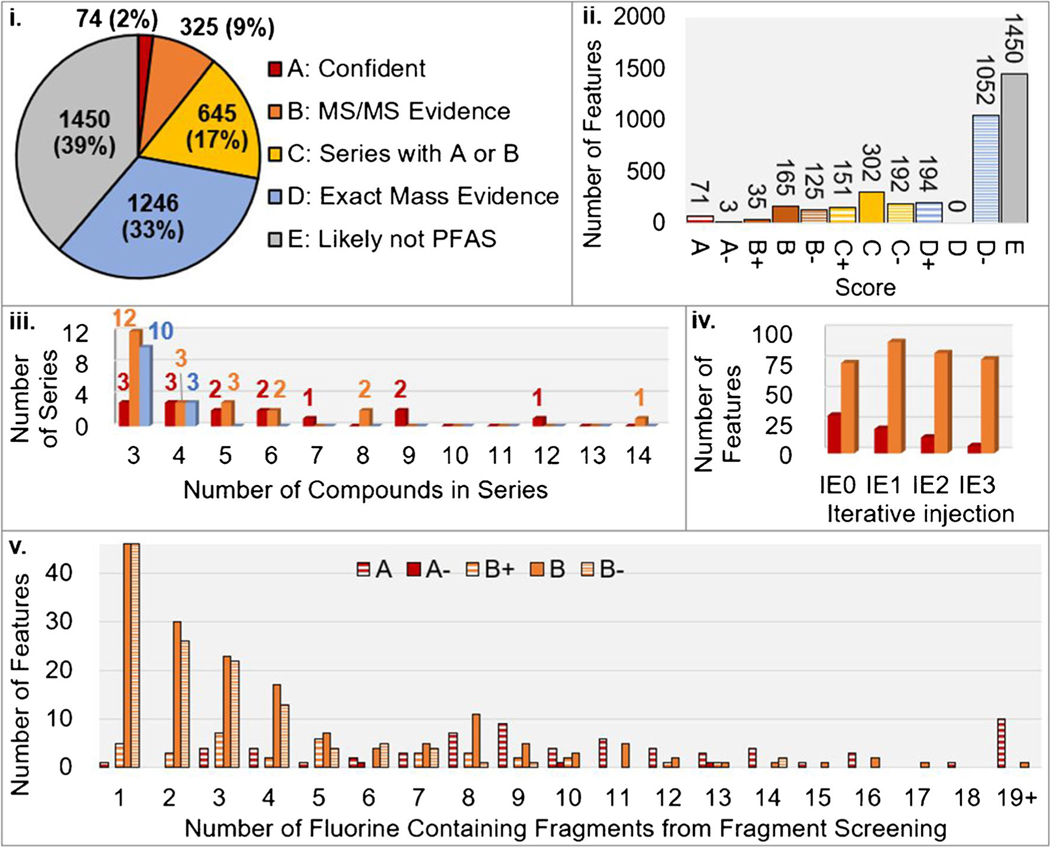
One advantage of FluoroMatch 2.0 is the automated reduction in features to only those which are likely PFAS for further manual validation. For the AFFF samples analyzed, a total of 31,132 features were detected. By implementing blank filtering in FluoroMatch 2.0, this feature list was reduced to 3740 features which were not from background (hence removing 88% of the detected features). Further reduction to likely PFAS features was achieved using our confidence scoring system. Of the blank-filtered features, 39% were deemed unlikely to be PFAS (score E), as they did not have any MS/MS evidence or exact mass evidence indicative of PFAS (Fig. 5i). These features assigned E are likely to be other constituents of AFFFs, for example hydrocarbon surfactants are more abundant than PFAS in AFFF [47]. Note that molecular formulae containing very few fluorine atoms and/or other atoms with a large impact on mass defect (e.g., iodine) may be in this “unlikely PFAS” (E) category. Of the remaining 2290 potential PFAS, 1238 formed homologous series and/or had at least one fluorine-containing fragment, and hence could be prioritized as probable PFAS (Fig. 5ii). The remaining 1052 (D−) had a mass defect within 98% of the PFAS mass defect range or an exact mass match, but without further evidence could likely be another chemical compound without fluorine. Hence, by using FluoroMatch, the feature list could easily be reduced by 96% to only those 1238 probable PFAS, drastically improving throughput in PFAS non-targeted analysis.
Fig. 5.
Overview of features proposed as potential PFAS, their scores, fragmentation evidence, and formation of homologous series (see Fig. 3 for definitions of scores). i The number and percent of features under each general scoring criteria and broad definition of scoring criteria (e.g., A is the sum of A+, A, and A−). ii Breakdown of the number of features across more refined scoring criteria A+, A, and A− (“−” for A and B mean the compounds were not observed within homologous series). iii Homologous series with 3 or more members; the top score for each series was used for classification by general scoring criteria for this figure (color codes are the same as in Fig. 5i). iv Number of features with new MS/MS assignments (score A or B) during iterative injections with a rolling exclusion list (color codes are the same as in Fig. 5i). v Number of fluorine-containing fragments using fragment screening (777 fragments screened) for features, categorized by score
After determining likely PFAS features, the next, and most time-consuming step when performed manually, is to assign structures to features. Using FluoroMatch 2.0, this step is automated and different levels of confidence are assigned to aid in interpretation (Fig. 3). Of the 1238 potential PFAS, MS/MS evidence was determined for 399 features (score A through B −) (Fig. 5ii). Without in-house retention time libraries (targeted analysis), MS/MS is needed for structural assignment of PFAS. Hence, improving MS/MS coverage during data acquisition is essential to increasing the number of FluoroMatch annotations.
Use of intelligent acquisition and homologous series detection to improve PFAS coverage
One difficulty is that mass spectra are feature rich, and yet only a small subset of ions can be isolated and fragmented using mass spectrometry in the time scale of liquid chromatography. Implementing iterative exclusion (IE) can drastically improve MS/MS coverage [30, 48]. In IE, an exclusion list is generated from ions selected for fragmentation in a first injection. In the following re-injections of the samples, a rolling exclusion list is applied to each iterative injection, excluding previously fragmented ions and fragmenting the next most abundant ions. Theoretically, this process can be repeated until all ions are fragmented. In the initial injection, only 105 features had MS/MS evidence for PFAS, whereas after 3 rounds of IE, 399 features had PFAS evidence (Fig. 5iv), hence increasing the possible PFAS which could be annotated by 280%. The number of confident annotations (score A) improved by 139% (more than doubling), with the number of additional confident annotations decreasing during subsequent iterative injections. This suggests that confident annotations were assigned to highly abundant compounds which could yield significant fragmentation evidence, and that nearly all high abundance compounds which could be assigned confident annotations were fragmented by the 4th injection (Fig. 5iv). Indeed, higher fragmentation coverage was observed for confident annotations (score A (A+, A, and A−) than for scores of B (B+, B, and B−) (Fig. 5v). Whereas further injections may not have improved confident annotation coverage, a significant number of additional features with some level of PFAS MS/MS evidence (score B) were still being obtained after the 4th injection (Fig. 5iv) and, hence, even further IE injections could be useful in improving the number of features with MS/MS evidence in this complex AFFF sample matrix.
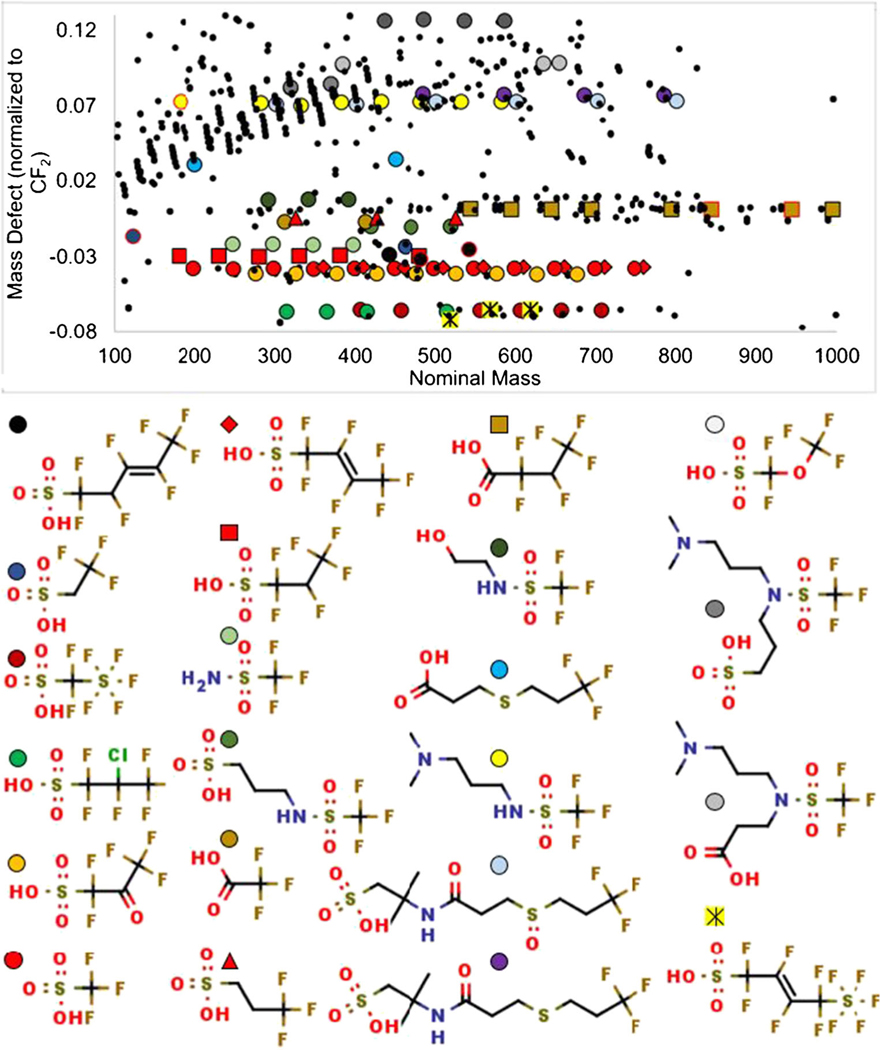
Even if MS/MS evidence cannot be obtained (for example, for low abundance compounds which do not yield indicative fragments above the noise level), possible structures can be tentatively assigned if other members of a homologous series are assigned a structure by MS/MS. In PFAS analysis, this can be achieved by plotting nominal mass against the CF2 normalized mass defect and searching for series with the same mass defect and nominal masses differing by intervals of 50 (CF2) (Fig. 6). Fifty homologous series were automatically detected by FluoroMatch 2.0 with 3 or more members (in this case defined as 3 or more unique CF2 chain lengths) (Fig. 5iii). Interestingly, series with 5 or more members all contained at least one feature with MS/MS evidence, with nearly 50% containing a member with a confident annotation. Hence, all major PFAS series detected had some evidence for structural assignment, and 50% had a confident structure assigned. Thirteen series, all with 3 to 4 members, had no MS/MS evidence.
Fig. 6.
Examples of PFAS homologous series automatically determined using FluoroMatch 2.0 with confident annotations (at least one score A: colored shapes), select tentative annotations (at least one score B: gray shapes), and newly discovered PFAS (yellow square with black lines). Red outlines are compounds flagged as tentative, for example, due to retention times not being in the correct order
Benchmarking FluoroMatch 2.0 against standards, traditional non-targeted approaches, and vendor algorithms
While it is clear FluoroMatch 2.0 can rapidly and comprehensively perform non-targeted PFAS analysis in an automated fashion, the question remains how confident the annotations provided are. Previous work has generally shown 100% agreement of score A and A− annotations with spiked standards, targeted analysis, and against other software platforms, with the exception of the inability to determine subtle differences including branched versus straight-chain isomers [30, 34]. In the present sample, 74 species were assigned confident annotations (A) using class-based fragmentation patterns. Furthermore, 251 species (including isomers) across 18 PFAS homologous series had at least one member of the series with a confident annotation (A) (Fig. 6) and hence had a confident starting structure for annotation.
This AFFF sample is also featured in previously published work [11], where PFAS were manually annotated, and we used this work as a comparison for our results (see ESM, Excel file). FluoroMatch 2.0 annotated 16 out of the 17 PFAS classes that were previously found using a manual non-targeted workflow. FluoroMatch 2.0 assigned identical, high confidence structures (level A) for 13 of these classes. This overlap increases confidence in FluoroMatch high-level annotations. The previous work assigned 50 species across their 17 annotated homologous series. FluoroMatch 2.0 assigned 87 species (not including branched isomers) for these same series. FluoroMatch 2.0 aided in the detection of a total of 22 PFAS series for which confident structures were assigned (Fig. 6), and 50 total homologous series, demonstrating a clear advantage in the number of annotations provided. A total of 75% of the FluoroMatch 2.0 series did not have confident annotations, showing further room for software improvements and the need for manual discovery.
Whereas FluoroMatch can be used to assign annotations from class-based standards and literature, one of the major advances of FluoroMatch 2.0 is the ability to predict fragmentation using substructure-fragment relationships (level B and B−). This enables MS/MS-based searching of any compound given a preset list of SMILES strings. A question remains regarding the confidence of these assignments. However, we found that only 5% (3 out of 58) of annotations based on in silico fragment masses differed from the A level structural assignments for features with both an in silico and level A assignment. Furthermore, two out of three FluoroMatch 2.0 structural annotations without level A assignments for comparison based on in silico fragment masses matched manually annotated PFAS series from the previous work. A third FluoroMatch in silico-based annotation differed from the previous work: FluoroMatch assigned (CN(C)CCCN(CCC(O) = O ) S ( = O ) ( = O ) C ( F ) ( F ) F … ) a n d ( C [ N + ](C)(CCCNS(=O)(=O)C(F)(F)F)CCCS(O)(=O) = O…) was derived manually. While this provides eveidence that in silico matches are confident, it was mainly the most common PFAS series for which the in silico approach was developed that were able to be benchmarked, and therefore further work on determining the confidence of this approach is warranted.
Therfore, further confirmation of level B and B− annotations at the level of formula was derived using Agilent MassHunter Qualitative Analysis 10.0 Find by Formula (FbF) compound mining algorithm. FbF automatically pulls extracted ion chromatograms (EICs) for all potential adducts and isotope signals in the MS level spectra, and bins them together as one compound entry. The extracted MS peak spectra are compared against the theoretical isotope distribution and scored based on accurate masses, the isotope pattern match, and the retention time, if stored in the target database. A target database containing formula, retention time, and the FluoroMatch 2.0 identifier for the relevant suspects was used as the FbF target formula source. Seventy-eight percent of the suspects with a FluoroMatch 2.0 B score were confidently identified (confirming formula), 11% were tentatively identified, and 11% were not found. For the suspects with a FluoroMatch 2.0 B− score, 33% of the suspects were confidently identified, 26% were tentatively identified, and 40% were not found. Upon inspection, the majority of the poor scoring suspects were found to be broad chromatographic peaks, many near the void volume. The one outlier was a series of level B compounds assigned as bromine containing PFAS. FbF proved these to be false positives, as the Brspecific isotopic pattern was not observed (ESM Fig. S3). Incorporating isotopic pattern matching in the MS level spectra is an opportunity for future improvement.
In summary, false positive rates for level A annotations are close to 0% based on previous work validating these assignments using authentic non-labeled standards, targeted workflows, and other algorithms [30, 34], and current work based on comparison to a manual non-targeted approach [11]. Hence, level A assignments can be trusted without thorough manual validation. Note, we are not considering structurally similar isomers such as branched versus linear chain isomers as false positives; these cannot be distinguished by fragmentation rules. Assignments using predicted fragmentation had a 5% false positive rate for this experiment, when they were annotations of a common PFAS which also had a level A annotation, but based on formula prediction, the actual rate of false positives is higher when accounting for all assignments. Hence, annotations and formula for level B annotations must be further investigated manually for confirmation. Therefore, we have added an additional column in the output labeled “Manual Validation Necessary” with a “yes” or “no” to ensure that users do not simply automatically use assignments which may be false positives.
Future investigations into false negatives (a compound present in the data, but not determined by the software) for level A and B assignments are necessary. Current applications suggest that peak picking and blank filtering are the major steps which can lead to false negatives, and the number and type of false negatives will greatly differ depending on users’ instrument and workflow. If the feature is picked, even if the software does not provide a PFAS annotation, the compound will likely be assigned as a C or D. Level C and D confidence assignments do not provide annotations, only evidence that the feature may be a PFAS, and in the case of C, may be a PFAS of the same series as other members with MS/MS evidence. This drastically reduces the chance of a complete false negative, a feature without any confidence assignment and PFAS-related evidence which actually is a PFAS. Further work examining false positives (assigned C or D, but not actually PFAS) and false negatives (actual PFAS but not assigned C or D) would be beneficial for C and D scoring criteria. For example, a sample void of PFAS could be used to determine the likelihood of false positive annotations.
Unknown-unknown discovery using FluoroMatch 2.0
Confident and predicted annotation using the EPA structure list, as well as automated determination of associated members of homologous series, can only determine PFAS structures which are “known unknowns” that is where their structures have been previously determined or hypothesized. One important aspect of non-targeted analysis is the detection of unknown-unknowns [49], that is chemicals which were previously unknown to exist or at least not found in accessible databases (e.g., trade secrets). Without knowledge of these unknowns, it is impossible to fully determine the scope of PFAS exposure and potential human health impacts. FluoroMatch 2.0 aids in the discovery of unknowns by reducing the potential features of interest to those which are likely PFAS (as previously discussed by 96% in this case) and assigning all confident annotations first. In this case, 36 of the 50 series automatically detected by FluoroMatch 2.0 with 3 or more members did not have a confident annotation. Of these compounds without database matches, 23 series had some MS/MS evidence and hence are good candidates for discovering new PFAS. Hence, a large number of distinct PFAS structures still remain to be discovered in AFFF.
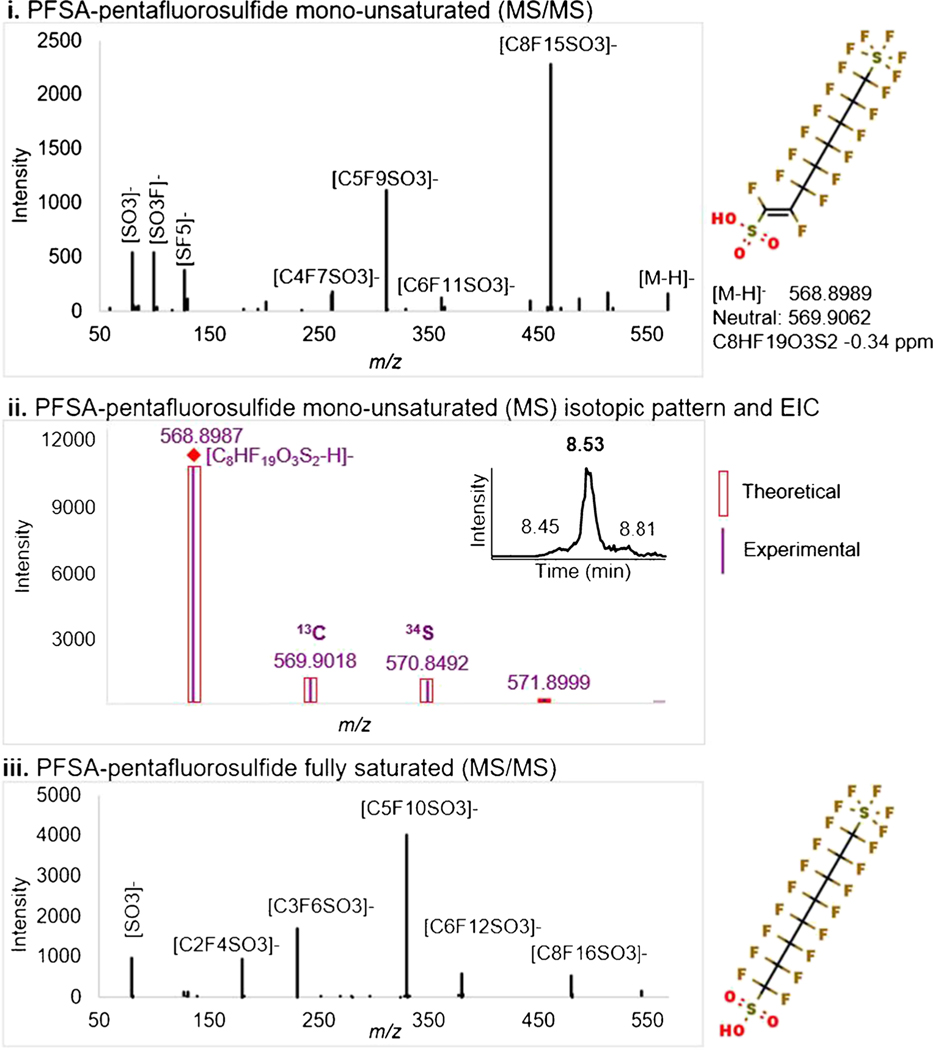
By using fragment screening, FluoroMatch 2.0 provides both fluorine-containing fragment assignments for unknowns and potential associated substructures for those fragments in SMILES form. This can greatly aid in manual discovery of unknowns. By using fragment screening, a new PFAS class was proposed in AFFF. This PFAS is proposed to contain pentafluorosulfide and a sulfonic acid terminus and one unsaturation (Fig. 7), with fully saturated versions previously identified. Fragment screening provided the following list of 12 fragments and potential substructures which may generate these fragments for the C8 member. Note that SMILES do not represent the potential fragment structure, but rather the substructure of the molecule which may generate this fragment, and these SMILES substructures are tentative:
Fig. 7.
Identification of unknown-unknowns (PFSA—pentafluorosulfide mono-unsaturated) two–three members detected using fragment screening. Comparison of a validated fully saturated species is shown for reference
From this list, it can be seen that the pentafluorosulfide and sulfonic acid substructures are correctly indicated through various fragments along with a CF2 chain of 8 units. The precursor mass is exactly two fluorine atoms lower than the fully saturated version, indicating one degree of unsaturation.
Furthermore, the unique isotopic patterns (due to S2) have a very high match between experimental and theoretical isotopic distribution (Fig. 7ii) as determined using Agilent’s find by formula algorithm. Fragmentation also was assigned using Agilent’s find by formula algorithm and matched FluoroMatch 2.0 annotations in nearly all cases (ESM Fig. S4). The only challenge to this interpretation is that the [C8F15SO3]− fragment (Fig. 7i), as well as others, has one additional F which would not be found in a monounsaturated chain. This suggests intermolecular transfer of a F during MS/MS. Fluorine transfer in MS/MS is extremely common, and loss of SF2 + F2, and retaining one F is possible. Note that the C10 member also provided similar fragmentation evidence, and that multiple isomers (like branched isomers) are possible based on chromatography (Fig. 7ii) and MS/MS evidence.
This discovery process could be implemented for the remaining unknowns with fragment screening annotations.
Hundreds of unknowns in the AFFF feature table have three or more assigned fluorine fragments, with tens of species which form homologous series having 8 or more assigned fragments (Fig. 5v). Hence, by manually scanning through assigned fragments and possible substructures associated with the fragments, the user is readily aided in discovering unknowns, and further work using the data provided in the ESM by the research community could further discover new PFAS in this legacy AFFF.
Conclusion and future developments
In an application with legacy AFFF samples, FluoroMatch Flow 2.0 increased coverage of likely PFAS by tenfold compared to the previous release. New capabilities responsible for this increase in coverage include homologous series detection, fragment screening, predicted MS/MS using substructure-fragment relationships, and a new scoring system for PFAS annotation confidence. The automated filtering to retain features which are likely PFAS drastically reduces the time spent on data analysis results review, in this case, reducing the number of features for review by 96%. Furthermore, a large portion of these compounds were assigned a confident annotation. Of the 50 homologous series automatically curated by FluoroMatch 2.0 with three or more members, nearly half (22) were confidently assigned a PFAS class using FluoroMatch 2.0.
In addition to providing confident structural assignments, as shown here and previously by using class-based fragmentation rules [30, 34], our algorithm provides a means for discovering new PFAS. Our new algorithms predicting fragmentation from SMILES structure had top hits matching high confidence annotations 95% of the time and were validated against previous traditional non-targeted workflows applied to these samples. It is important to note, false positive rates increase for these structural assignments from in silico fragmentation for less common or abundant PFAS as shown by comparison to Agilent’s find by formula algorithm. In addition to in silico fragmentation improving coverage, fragment screening provided a wealth of information for over 350 features for the discovery of unknown PFAS. As an example, we show the tentative identification of a previously unreported unsaturated PFAS with a pentafluorosulfide and sulfonic acid termini.
FluoroMatch 2.0 will drastically increase the throughput of researchers performing non-targeted PFAS analysis and improve PFAS annotation coverage. Furthermore, our in-depth scoring rubric provides a platform for harmonization of PFAS-targeted, non-targeted, and suspect screening data reporting. By integrating this scoring rubric in FluoroMatch 2.0, this harmonized method of reporting data is more likely to be adopted.
Future improvements to FluoroMatch 2.0 compound coverage mainly relate to library expansion, with a user workflow available for community-driven expansion of libraries. Currently libraries contain only [M-H]− ions, but FluoroMatch 2.0 supports various adducts and positive polarity, and therefore expansion of FluoroMatch libraries beyond a single type of adduct could further improve coverage. Finally, accurate formula prediction is not currently available in R to our knowledge, with packages we have tested failing with F-containing compounds, and therefore accurate and efficient formula prediction in R would be a powerful additional tool to integrate into FluoroMatch.
While assignments of A have been found to be confident, the false positive rates of other assignment need to be fully investigated, in addition to the false negative rates of all assignments. Further algorithms to reduce false positives may be developed in the future. While libraries contain formulas which theoretical isotopic patterns can be derived from, isotopic pattern matching is currently not implemented and will reduce false positives as shown in this study. In addition to isotopic pattern, intensity information in fragmentation is not accounted for, which would further reduce false positives. This information is challenging to accurately simulate across the diverse range of PFAS species, especially given the limited number of standards available to develop a training set. Predicted retention times would also drastically reduce false positives, but the complex chemistries of liquid chromatography also make this highly challenging.
Supplementary Material
Acknowledgements
A large array of PFAS standards used to develop libraries were donated by SynQuest Labs, Inc. and Oakwood Products, Inc.
Funding
S.L.N was supported by USDA NIFA Hatch funds (CONH00789). JAB received support from the U.S. Environmental Protection Agency under the Science To Achieve Results (STAR) grant programs: EPA-G2018-STAR-B1-Grant#: 83962001-0-and EPA-G2019-STAR-E1-Grant#: 84004501-0. J.P.K and K.J.G.P. received support from Agilent Technologies ACT-UR grant mechanism.
Footnotes
Data availability The final annotated excel sheets with feature intensities, annotations, homologous series groupings, etc., are available as a supplemental excel file with the online version of this manuscript. The raw Agilent “.d” files can be downloaded at: ftp://massive.ucsd.edu/MSV000086811/updates/2021–02-05_jeremykoelmel_e5b21166/raw/McDonough_AFFF_3M_ddMS2_Neg.zip
(Note use Google Chrome or Firefox, Microsoft Edge and certain other browsers are unable to download from an ftp link).
Code availability The FluoroMatch Software platform and written and video tutorials are available at: http://innovativeomics.com/software/fluoromatch-flow-covers-entire-pfas-workflow/
Declarations
Competing interests The authors declare no competing interests.
Supplementary Information The online version contains supplementary material available at https://doi.org/10.1007/s00216-021-03392-7.
References
- 1.EUR-Lex - 32017R1000 - EN - EUR-Lex. https://eur-lex.europa.eu/eli/reg/2017/1000/oj. Accessed 30 Oct 2020.
- 2.Synthesis paper on per and polyfluorinated chemicals - OECD. https://www.oecd.org/chemicalsafety/risk-management/synthesis-paper-on-per-and-polyfluorinated-chemicals.htm. Accessed 30 Oct 2020.
- 3.Glüge J, Scheringer M, Cousins TI, DeWitt CJ, Goldenman G, Herzke D, et al. An overview of the uses of per- and polyfluoroalkyl substances (PFAS). Environ Sci Process Impacts. 2020;22:2345–73. 10.1039/D0EM00291G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Filipovic M, Woldegiorgis A, Norström K, Bibi M, Lindberg M, Österås A-H. Historical usage of aqueous film forming foam: a case study of the widespread distribution of perfluoroalkyl acids from a military airport to groundwater, lakes, soils and fish. Chemosphere. 2015;129:39–45. 10.1016/j.chemosphere.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 5.Hu XC, Andrews DQ, Lindstrom AB, Bruton TA, Schaider LA, Grandjean P, et al. Detection of poly- and perfluoroalkyl substances (PFASs) in U.S. drinking water linked to industrial sites, military fire training areas, and wastewater treatment plants. Environ Sci Technol Lett. 2016;3:344–50. 10.1021/acs.estlett.6b00260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Langberg HA, Breedveld GD, Grønning HM, Kvennås M, Jenssen BM, Hale SE. Bioaccumulation of fluorotelomer sulfonates and perfluoroalkyl acids in marine organisms living in aqueous film-forming foam impacted waters. Environ Sci Technol. 2019;53: 10951–60. 10.1021/acs.est.9b00927. [DOI] [PubMed] [Google Scholar]
- 7.Zhao S, Zhu L, Liu L, Liu Z, Zhang Y. Bioaccumulation of perfluoroalkyl carboxylates (PFCAs) and perfluoroalkane sulfonates (PFSAs) by earthworms (Eisenia fetida) in soil. Environ Pollut. 2013;179:45–52. 10.1016/j.envpol.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 8.Martín J, Hidalgo F, García-Corcoles MT, Ibáñez-Yuste AJ, Alonso E, Vilchez JL, et al. Bioaccumulation of perfluoroalkyl substances in marine echinoderms: results of laboratory-scale experiments with Holothuria tubulosa Gmelin, 1791. Chemosphere. 2019;215:261–71. 10.1016/j.chemosphere.2018.10.037. [DOI] [PubMed] [Google Scholar]
- 9.Haukås M, Berger U, Hop H, Gulliksen B, Gabrielsen GW. Bioaccumulation of per- and polyfluorinated alkyl substances (PFAS) in selected species from the Barents Sea food web. Environ Pollut. 2007;148:360–71. 10.1016/j.envpol.2006.09.021. [DOI] [PubMed] [Google Scholar]
- 10.Munoz G, Desrosiers M, Vetter L, Vo Duy S, Jarjour J, Liu J, et al. Bioaccumulation of zwitterionic polyfluoroalkyl substances in earthworms exposed to aqueous film-forming foam impacted soils. Environ Sci Technol. 2020;54:1687–97. 10.1021/acs.est.9b05102. [DOI] [PubMed] [Google Scholar]
- 11.McDonough CA, Choyke S, Ferguson PL, DeWitt JC, Higgins CP. Bioaccumulation of novel per- and polyfluoroalkyl substances in mice dosed with an aqueous film-forming foam. Environ Sci Technol. 2020;54:5700–9. 10.1021/acs.est.0c00234. [DOI] [PubMed] [Google Scholar]
- 12.Sunderland EM, Hu XC, Dassuncao C, Tokranov AK, Wagner CC, Allen JG. A review of the pathways of human exposure to poly- and perfluoroalkyl substances (PFASs) and present understanding of health effects. J Expos Sci Environ Epidemiol. 2019;29:131–47. 10.1038/s41370-018-0094-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fenton SE, Ducatman A, Boobis A, DeWitt JC, Lau C, Ng C, et al. Per- and polyfluoroalkyl substance toxicity and human health review: current state of knowledge and strategies for informing future research. Environ Toxicol Chem. 2020. 10.1002/etc.4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nelson JW, Hatch EE, Webster TF. Exposure to polyfluoroalkyl chemicals and cholesterol, body weight, and insulin resistance in the general U.S. population. Environ Health Perspect. 2010;118: 197–202. 10.1289/ehp.0901165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lopez-Espinosa M-J, Mondal D, Armstrong B, Bloom MS, Fletcher T. Thyroid function and perfluoroalkyl acids in children living near a chemical plant. Environ Health Perspect. 2012;120: 1036–41. 10.1289/ehp.1104370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Steenland K, Tinker S, Frisbee S, Ducatman A, Vaccarino V. Association of perfluorooctanoic acid and perfluorooctane sulfonate with serum lipids among adults living near a chemical plant. Am J Epidemiol. 2009;170:1268–78. 10.1093/aje/kwp279. [DOI] [PubMed] [Google Scholar]
- 17.Olsen GW, Burris JM, Burlew MM, Mandel JH. Epidemiologic assessment of worker serum perfluorooctanesulfonate (PFOS) and perfluorooctanoate (PFOA) concentrations and medical surveillance examinations. J Occup Environ Med. 2003;45:260–70. 10.1097/01.jom.0000052958.59271.10. [DOI] [PubMed] [Google Scholar]
- 18.Darrow LA, Stein CR, Steenland K. Serum perfluorooctanoic acid and perfluorooctane sulfonate concentrations in relation to birth outcomes in the Mid-Ohio Valley, 2005–2010. Environ Health Perspect. 2013;121:1207–13. 10.1289/ehp.1206372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Szilagyi JT, Avula V, Fry RC. Perfluoroalkyl substances (PFAS) and their effects on the placenta, pregnancy, and child development: a potential mechanistic role for placental peroxisome proliferator–activated receptors (PPARs). Curr Environ Health Rpt. 2020;7: 222–30. 10.1007/s40572-020-00279-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Steenland K, Zhao L, Winquist A, Parks C. Ulcerative colitis and perfluorooctanoic acid (PFOA) in a highly exposed population of community residents and workers in the mid-Ohio valley. Environ Health Perspect. 2013;121:900–5. 10.1289/ehp.1206449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barry V, Winquist A, Steenland K. Perfluorooctanoic acid (PFOA) exposures and incident cancers among adults living near a chemical plant. Environ Health Perspect. 2013;121:1313–8. 10.1289/ehp.1306615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shearer JJ, Callahan CL, Calafat AM, Huang W-Y, Jones RR, Sabbisetti VS, et al. Serum concentrations of per- and polyfluoroalkyl substances and risk of renal cell carcinoma. J Natl Cancer Inst. 10.1093/jnci/djaa143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grandjean P, Heilmann C, Weihe P, Nielsen F, Mogensen UB, Timmermann A, et al. Estimated exposures to perfluorinated compounds in infancy predict attenuated vaccine antibody concentrations at age 5-years. J Immunotoxicol. 2017;14:188–95. 10.1080/1547691X.2017.1360968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DeWitt JC, Peden-Adams MM, Keller JM, Germolec DR. Immunotoxicity of perfluorinated compounds: recent developments. Toxicol Pathol. 2012;40:300–11. 10.1177/0192623311428473. [DOI] [PubMed] [Google Scholar]
- 25.McDonough CA, Ward C, Hu Q, Vance S, Higgins CP, DeWitt JC. Immunotoxicity of an electrochemically fluorinated aqueous film-forming foam. Toxicol Sci. 2020;178:104–14. 10.1093/toxsci/kfaa138. [DOI] [PubMed] [Google Scholar]
- 26.Yeung LWY, Miyake Y, Taniyasu S, Wang Y, Yu H, So MK, et al. Perfluorinated compounds and total and extractable organic fluorine in human blood samples from China. Environ Sci Technol. 2008;42:8140–5. 10.1021/es800631n. [DOI] [PubMed] [Google Scholar]
- 27.Goodrum PE, Anderson JK, Luz AL, Ansell GK. Application of a framework for grouping and mixtures toxicity assessment of PFAS: a closer examination of dose-additivity approaches. Toxicol Sci. 10.1093/toxsci/kfaa123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.CompTox Chemicals Dashboard | PFASSTRUCT Chemicals. https://comptox.epa.gov/dashboard/chemical_lists/PFASSTRUCT. Accessed 22 Jan 2021.
- 29.Williams AJ, Grulke CM, Edwards J, McEachran AD, Mansouri K, Baker NC, et al. The CompTox Chemistry Dashboard: a community data resource for environmental chemistry. J Cheminform. 2017;9:61. 10.1186/s13321-017-0247-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koelmel JP, Paige MK, Aristizabal-Henao JJ, Robey NM, Nason SL, Stelben PJ, et al. Toward comprehensive per- and polyfluoroalkyl substances annotation using FluoroMatch software and intelligent high-resolution tandem mass spectrometry acquisition. Anal Chem. 2020;92:11186–94. 10.1021/acs.analchem.0c01591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McDonough CA, Guelfo JL, Higgins CP. Measuring total PFASs in water: the tradeoff between selectivity and inclusivity. Curr Opin Environ Sci Health. 2019;7:13–8. 10.1016/j.coesh.2018.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Y, D’Agostino L, Qu G, Jiang G, Martin J. High-resolution mass spectrometry (HRMS) methods for nontarget discovery and characterization of poly- and per-fluoroalkyl substances (PFASs) in environmental and human samples. TrAC Trends Anal Chem. 2019. 10.1016/j.trac.2019.02.021. [DOI] [Google Scholar]
- 33.Bugsel B, Zwiener C. LC-MS screening of poly- and perfluoroalkyl substances in contaminated soil by Kendrick mass analysis. Anal Bioanal Chem. 2020;412:4797–805. 10.1007/s00216-019-02358-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nason SL, Koelmel J, Zuverza-Mena N, Stanley C, Tamez C, Bowden JA, et al. Software comparison for nontargeted analysis of PFAS in AFFF-contaminated soil. J Am Soc Mass Spectrom. 2019. 10.1021/jasms.0c00261. [DOI] [PubMed] [Google Scholar]
- 35.D’Agostino LA, Mabury SA. Identification of novel fluorinated surfactants in aqueous film forming foams and commercial surfactant concentrates. Environ Sci Technol. 2014;48:121–9. 10.1021/es403729e. [DOI] [PubMed] [Google Scholar]
- 36.Place BJ, Field JA. Identification of novel fluorochemicals in aqueous film-forming foams used by the US military. Environ Sci Technol. 2012;46:7120–7. 10.1021/es301465n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barzen-Hanson KA, Roberts SC, Choyke S, Oetjen K, McAlees A, Riddell N, et al. Discovery of 40 classes of per- and polyfluoroalkyl substances in historical aqueous film-forming foams (AFFFs) and AFFF-impacted groundwater. Environ Sci Technol. 2017;51:2047–57. 10.1021/acs.est.6b05843. [DOI] [PubMed] [Google Scholar]
- 38.Awad E, Zhang X, Bhavsar SP, Petro S, Crozier PW, Reiner EJ, et al. Long-term environmental fate of perfluorinated compounds after accidental release at Toronto airport. Environ Sci Technol. 2011;45:8081–9. 10.1021/es2001985. [DOI] [PubMed] [Google Scholar]
- 39.Oakes KD, Benskin JP, Martin JW, Ings JS, Heinrichs JY, Dixon DG, et al. Biomonitoring of perfluorochemicals and toxicity to the downstream fish community of Etobicoke Creek following deployment of aqueous film-forming foam. Aquat Toxicol. 2010;98:120–9. 10.1016/j.aquatox.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 40.Moody CA, Martin JW, Kwan WC, Muir DCG, Mabury SA. Monitoring perfluorinated surfactants in biota and surface water samples following an accidental release of fire-fighting foam into Etobicoke Creek. Environ Sci Technol. 2002;36:545–51. 10.1021/es011001+. [DOI] [PubMed] [Google Scholar]
- 41.Guelfo JL, Adamson DT. Evaluation of a national data set for insights into sources, composition, and concentrations of per- and polyfluoroalkyl substances (PFASs) in U.S. drinking water. Environ Pollut. 2018;236:505–13. 10.1016/j.envpol.2018.01.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mueller R, Yingling V. Aqueous film-forming foam (AFFF), Interstate Technol Regul Counc Sheets (ITRC). 2018. [Google Scholar]
- 43.Anderson RH, Thompson T, Stroo HF, Leeson A. US Department of Defense-funded fate and transport research on per- and polyfluoroalkyl substances at aqueous film-forming foam-impacted sites. Environ Toxicol Chem. 2021;40:37–43. 10.1002/etc.4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Draper J, Enot DP, Parker D, Beckmann M, Snowdon S, Lin W, et al. Metabolite signal identification in accurate mass metabolomics data with MZedDB, an interactive m/z annotation tool utilising predicted ionisation behaviour “rules”. BMC Bioinform. 2009;10:227. 10.1186/1471-2105-10-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guha R Chemical informatics functionality in R. J Stat Softw. 2007;18. [Google Scholar]
- 46.Schymanski EL, Jeon J, Gulde R, Fenner K, Ruff M, Singer HP, et al. Identifying small molecules via high resolution mass spectrometry: communicating confidence. Environ Sci Technol. 2014;48:2097–8. 10.1021/es5002105. [DOI] [PubMed] [Google Scholar]
- 47.García RA, Chiaia-Hernández AC, Lara-Martin PA, Loos M, Hollender J, Oetjen K, et al. Suspect screening of hydrocarbon surfactants in AFFFs and AFFF-contaminated groundwater by high-resolution mass spectrometry. Environ Sci Technol. 2019;53:8068–77. 10.1021/acs.est.9b01895. [DOI] [PubMed] [Google Scholar]
- 48.Koelmel JP, Kroeger NM, Gill EL, Ulmer CZ, Bowden JA, Patterson RE, et al. Expanding lipidome coverage using LC-MS/MS data-dependent acquisition with automated exclusion list generation. J Am Soc Mass Spectrom. 2017;28:908–17. 10.1007/s13361-017-1608-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Little JL, Williams AJ, Pshenichnov A, Tkachenko V. Identification of “known unknowns” utilizing accurate mass data and ChemSpider. J Am Soc Mass Spectrom. 2012;23:179–85. 10.1007/s13361-011-0265-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.