Figure 4.
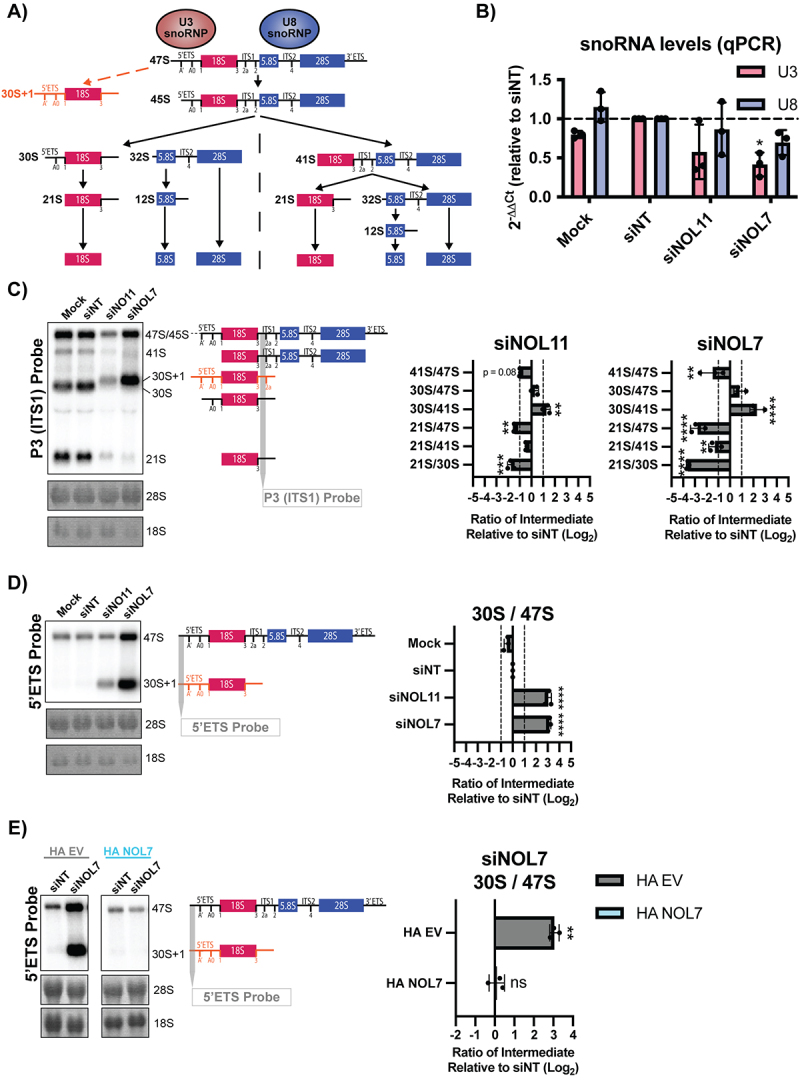
NOL7 is required for U3 snoRNA-mediated small subunit pre-rRNA processing in human tissue culture cells.
A. pre-rRNA processing in human cells. The 47S pre-rRNA precursor undergoes a series of modification and processing steps to yield the mature 18S, 5.8S, and 28S rRNAs. Upon inhibition of pre-rRNA cleavage in the 5’ETS, mediated by the U3 snoRNP, accumulation of an 5’ETS extended 30S + 1 precursor occurs (orange). The steps that produce the 18S rRNA (small subunit) are indicated in red, and the steps that produce the 5.8S and 28S rRNAs (large subunit) are indicated in blue. The U8 snoRNA is essential for processing of the rRNAs that make the large ribosomal subunit.
B. NOL7 siRNA depletion in MCF10A cells results in reduced U3 snoRNA steady-state levels. qRT-PCR measuring U3 and U8 snoRNA transcript levels in MCF10A cells. 2−ΔΔCt were measured relative to 7SL internal control and siNT negative control sample. 3 technical replicates of 3 biological replicates, plotted mean ± SD, dotted line at siNT value y = 1. Data were analysed by one-way ANOVA with Dunnett’s multiple comparisons test, * p ≤ 0.05.
C. NOL7 is required for pre-18S rRNA processing in MCF10A cells. (Left) Representative northern blot using a P3 ITS1 probe (indicated in grey) measuring steady state levels of pre-rRNA precursors leading to the 18S rRNA. The 47S/45S precursors are not separated on these blots but are both detected with this probe. Methylene blue staining of 28S and 18S was used to show even loading. siNT is a negative control and siNOL11 is a positive control. (Right) Quantification of northern blots using ratio analysis of multiple precursors (RAMP) 15 relative to siNT negative control. 3 biological replicates, plotted mean ± SD. Data were analysed by two-way ANOVA, ** p ≤ 0.01, *** p ≤ 0.001, **** ≤ 0.0001. The 47S precursor ratios on this graph are a combination of the quantification of the indicated 47S/45S band (left) since these precursors are not separated on these blots.
D. NOL7 is required for 5’ETS cleavage in MCF10A cells. (Left) Representative northern blot using a 5’ETS probe (upstream of the A’ site, indicated in grey) measuring steady state levels of 47S and 30S + 1 pre-rRNA precursors. Methylene blue staining of 28S and 18S was used to show even loading. siNT is a negative control and siNOL11 is a positive control. (Right) Quantification of northern blots using ratio analysis of multiple precursors (RAMP) 15 relative to siNT negative control. 3 biological replicates, plotted Log2 (mean ± SD). Data were analysed by one-way ANOVA with Dunnett’s multiple comparisons test, **** p ≤ 0.0001.
E. Accumulation of the 30S + 1 pre-rRNA after siNOL7 depletion can be rescued by introduction of an si-resistant version of NOL7 in Hela cells. (Left) Representative northern blot using a 5’ETS probe (upstream of the A’ site, indicated in grey) measuring steady state levels of 47S and 30S + 1 pre-rRNA precursors upon NOL7 depletion and si-resistant rescue with either empty vector (HA EV) or HA-tagged NOL7 (HA NOL7) in HeLa cells. Methylene blue staining of 28S and 18S show even loading. siNT is a negative control. (Right) Quantification of northern blots using ratio analysis of multiple precursors (RAMP) 15 relative to siNT negative control. 3 biological replicates, plotted Log2 (mean ± SD). Data were analysed by Student’s t-test, ** p ≤ 0.05, ns = not significant.
