Figure 1: Active promoters/TSSs form CTCF-independent chromatin domain boundaries.
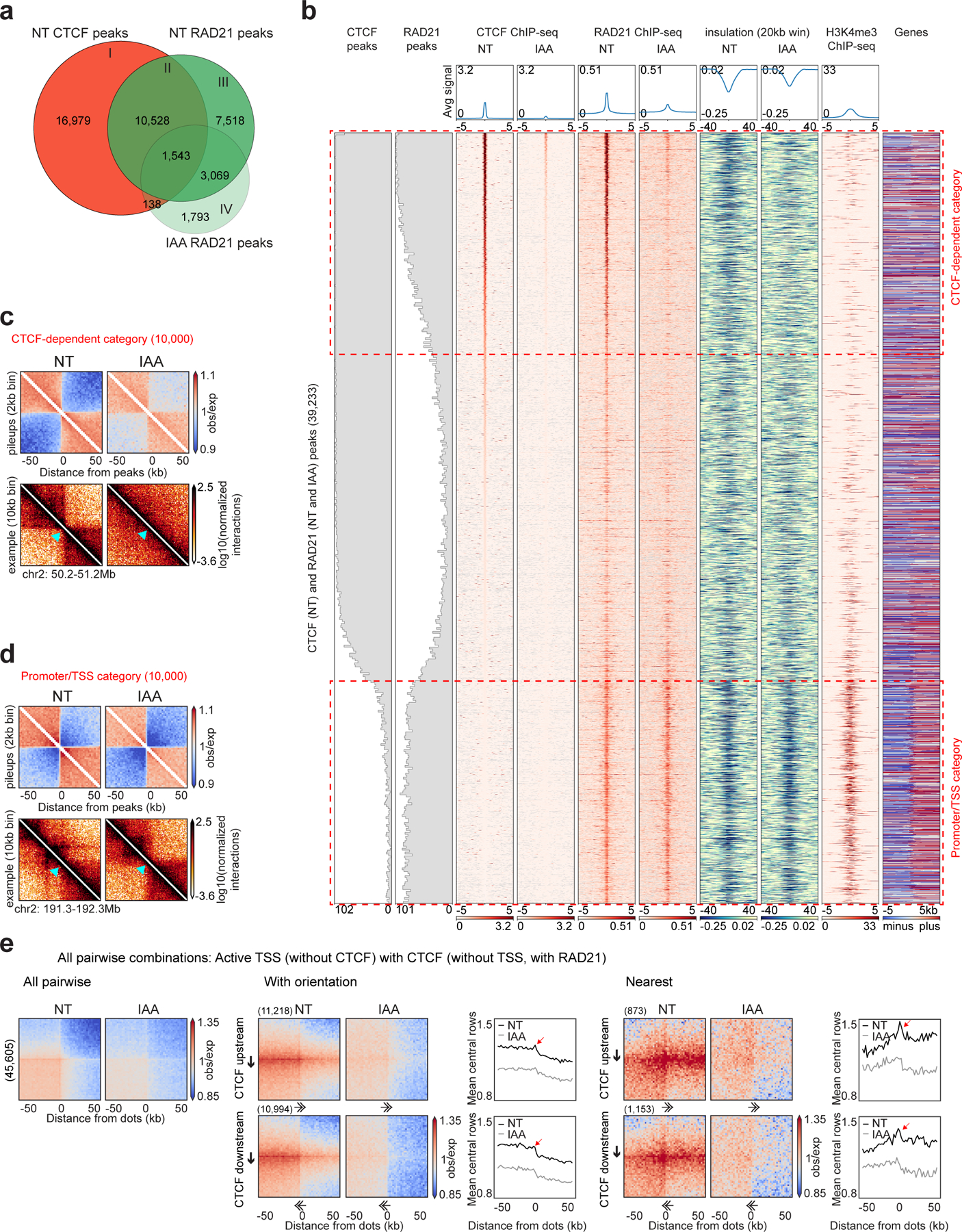
a, Venn diagram showing the overlap between CTCF peaks without auxin (NT), RAD21 peaks without/with auxin (NT and IAA).
b, Stackups of the union list of all CTCF peaks called without auxin and RAD21 peaks called without/with auxin, sorted on NT CTCF ChIP-seq signal. CTCF and RAD21 ChIP-seq and calculated insulation without/with auxin were plotted along with the published HAP1 H3K4me3 ChIP-seq and the genes (red, plus: forward strand, blue, minus: reverse strand, grey: no annotated transcripts)26. The distribution of CTCF and RAD21 peaks were plotted along the stackup. Red dashed rectangles highlight CTCF-dependent category and promoter/TSS category.
c, Interaction pileup for the CTCF-dependent category (10,000 sites), without/with auxin, aggregated in a 100kb window at 2kb resolution and plotted along with a representative example of a boundary. The light blue arrow shows the boundary location.
d, Interaction pileup for the promoter/TSS category (10,000 sites), as described in 1c.
e, Dot pileup aggregation plots for pairwise combinations of active TSSs (without CTCF) and CTCF peaks (without TSSs, with RAD21) separated by 50–500kb, without/with auxin, for a 100kb window at 2kb resolution. All pairwise interactions (left). With orientation (middle). CTCF (upstream or downstream)-TSS pairwise interactions are plotted with their quantification (mean of the 5 central bins at the CTCF site). Nearest analysis (right). CTCF (upstream or downstream)-TSS pairwise interactions are plotted without any CTCF peaks or TSSs in between them with their quantification (mean of the 5 central bins at the CTCF site). The red arrows represent the peak of interactions between CTCF site and TSS. The black arrows represent the CTCF motif and the direction of the arrow, the motif orientation. The double arrows represent the TSS and the direction of the arrow, the TSS orientation.
