Extended Data Fig. 9. Characterization of DDX55 and TAF5L depletions in HAP1-CTCFdegron-TIR1 cells.
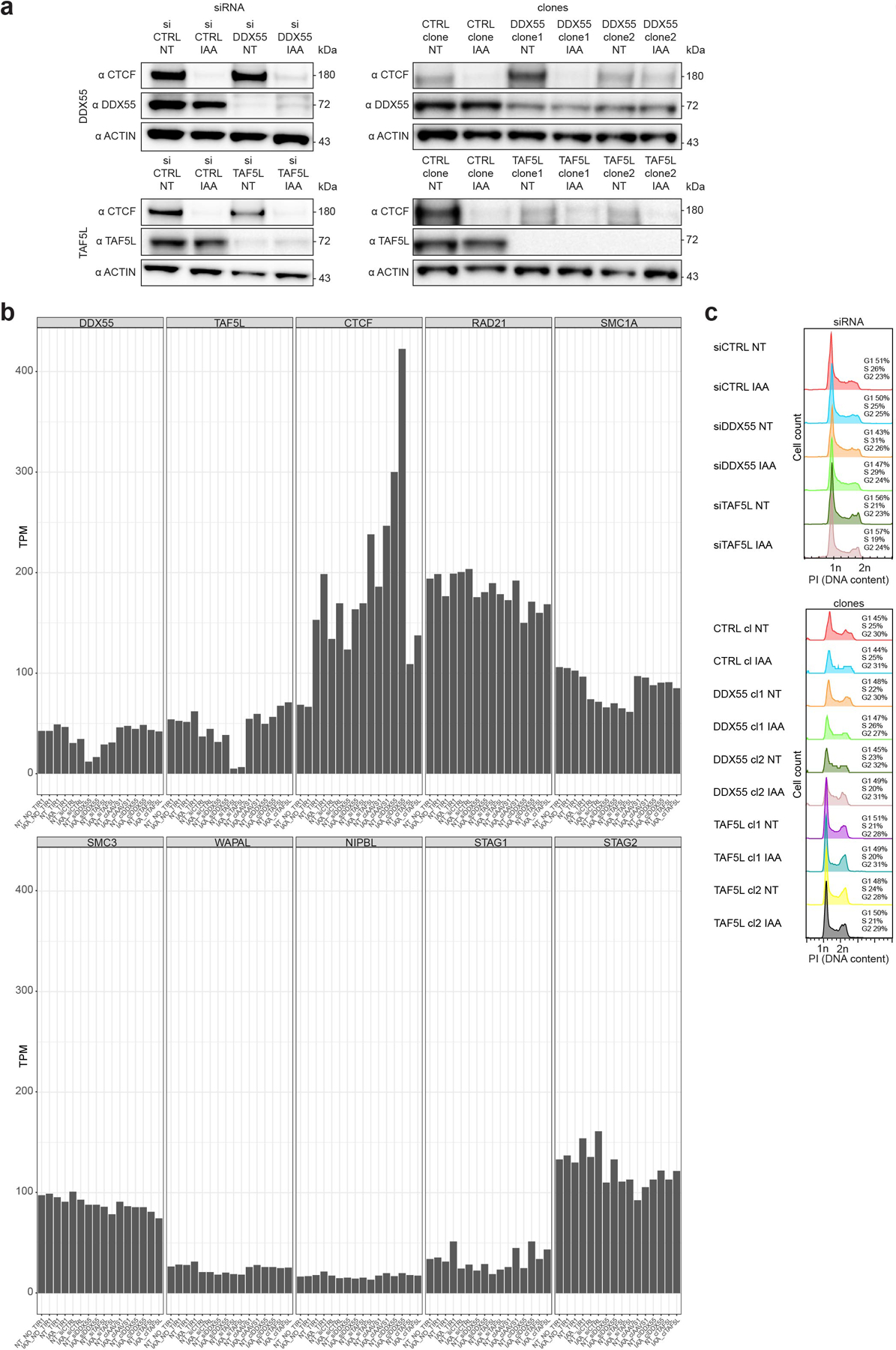
a, Western blots against CTCF, DDX55, TAF5L and β-ACTIN (loading control) showing DDX55 and TAF5L depletions, by siRNA and mutations in DDX55 and TAF5L genes, compared to siRNA controls and mutations at the AAVS1 non-coding sequence (CTRL clone).
b, RNA-seq expression (TPM) for HAP1-CTCFdegron-TIR1 cells without/with auxin (NT and IAA) treated with siRNA (siCTRL, siDDX55 and siTAF5L) and CTRL, DDX55 and TAF5L clones for key genes (DDX55, TAF5L, CTCF, RAD21, SMC1A, SMC3, WAPAL, NIPBL, STAG1 and STAG2).
c, Flow cytometry for HAP1-CTCFdegron-TIR1 cells without/with auxin (NT and IAA) with siRNA (siCTRL (control), siDDX55 and siTAF5L) (top) and CTRL (control), DDX55 and TAF5L clones (bottom) stained with Propidium Iodide (PI) to assess the DNA content for cell cycle analysis.
