Figure 5: Altered cohesin traffic pattern following CTCF depletion reduces chromatin binding of DDX55 and TAF5L at active promoters/TSSs.
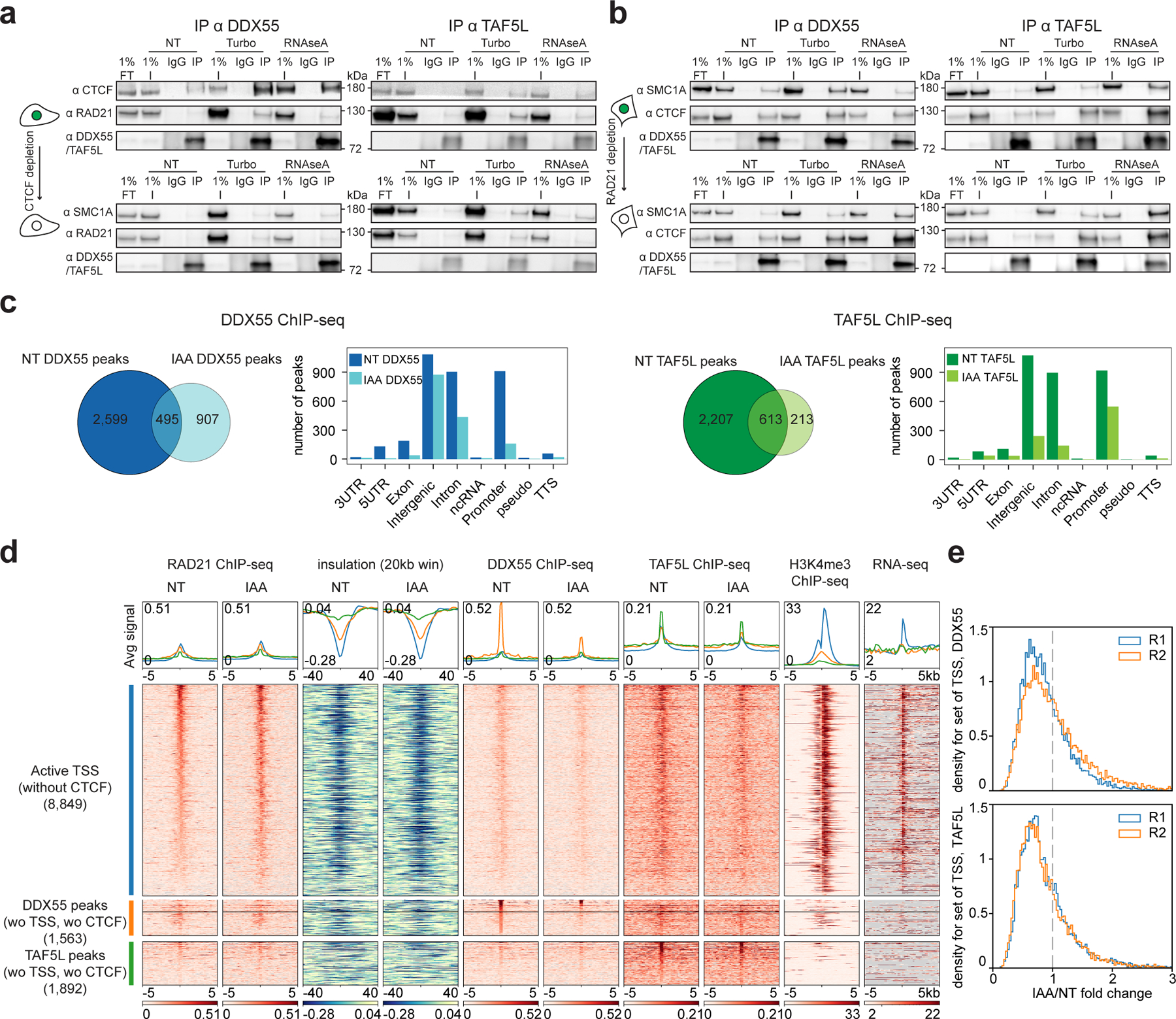
a, Western blot co-IPs against DDX55 and TAF5L in HAP1-CTCFdegron-TIR1 without/with auxin (NT and IAA), treated with either turbonuclease (DNA - and RNA -) or RNAseA (RNA -).
b, Western blot co-IPs against DDX55 and TAF5L in RAD21-AID degron without/with auxin (NT and IAA), treated with either turbonuclease (DNA - and RNA -) or RNAseA (RNA -).
c, Venn diagram for DDX55 (left) and TAF5L (right) ChIP-seq peaks called in HAP1-CTCFdegron-TIR1 cells without/with auxin (NT and IAA). Gene annotation bar plots of the ChIP-seq peaks called in HAP1-CTCFdegron-TIR1 without/with auxin (NT and IAA).
d, Stackups for active TSSs without CTCF (blue) sorted on Non-Treated (NT) RAD21 ChIP-seq signal, for DDX55 peaks (without TSSs, without CTCF) (orange) sorted on Non-Treated (NT) DDX55 ChIP-seq, and for TAF5L peaks (without TSSs, without CTCF) (green) sorted on Non-Treated (NT) TAF5L ChIP-seq. RAD21 ChIP-seq, calculated insulation, DDX55 ChIP-seq, TAF5L ChIP-seq and RNA seq signals in HAP1-CTCFdegron-TIR1 cells without/with auxin (NT and IAA) were plotted along with published HAP1 H3K4me3 ChIP-seq26 and oriented genes. For TSSs, genes were flipped according to their orientations to have the gene body on the right of the TSSs. Genes were not flipped for DDX55 and TAF5L peaks.
e, Stackup quantification for the active TSSs (without CTCF) for DDX55 and TAF5L ChIP-seq for two replicates. The distribution of the ratios (fold change) of a given signal between auxin-treated and non-treated conditions is shown. A fold change < 1 represents less binding of DDX55 or TAF5L at active TSSs after CTCF depletion.
