Abstract
Objective:
The rectal mucosa is a critical site of HIV vulnerability. We sought to identify transcriptomic features of rectal mucosal tissue prior to exposure associated with support or restriction of HIV replication.
Design:
Rectal tissue from HIV-negative cisgender men (n=57) underwent concurrent i) RNAseq transcriptomic analyses (2 biopsies/participant) and ii) challenge with HIV in the ex vivo explant model of infection (3 biopsies challenged/participant) as part of a larger cohort study to understand the rectal mucosal immune environment among men who have sex with men.
Methods:
P24 was quantified in the explant supernatants over a culture period of 18 days via ELISA. Participant median p24 log Area Under the Curve was correlated with bulk transcriptomic data (Illumina HiSeq3000) to identify associations between gene expression and p24 production. Significant differentially expressed genes (DEG) were identified via DESeq2 analysis, and analyzed with Reactome to identify pathways of interest.
Results:
183 DEG (181 upregulated, 2 downregulated) were associated with higher p24 accumulation in the ex vivo challenge model, including T cell activation, B cell function, and chemokine DEG. Reactome analysis of the upregulated genes identified ‘Adaptive Immune System’, ‘Cytokine Signaling in Immune System’, and ‘Innate Immune System’ as significant upregulated pathways.
Conclusions:
For the first time, we identified rectal tissue transcriptomic signatures associated with increased p24 production utilizing an ex vivo model. Our findings are highly relevant to HIV transmission and the early establishment of HIV reservoirs in humans, and future studies should examine the identified pathways as targets for new or improved biomedical prevention or treatment interventions.
Introduction
Among sexual exposure routes, the highest risk of HIV acquisition is through receptive anal intercourse,[1] contributing to a substantial number of new HIV diagnoses in men who have sex with men (MSM), as well as an unknown percentage of heterosexual individuals.[2–6] During rectal HIV transmission, mucosal microtears or transcytosis through the epithelial barrier facilitates HIV access to an abundance of target CD4+ T cells within the gut.[7] However, the gut is a diverse ecosystem of molecules, cells, and microbes, each of which could influence HIV replication within this tissue compartment. While a vaccine to prevent HIV transmission remains a top priority, a better understanding of the biological signatures contributing to or restricting HIV replication within gut tissue at the time of HIV exposure could provide critical complementary information for ongoing vaccination efforts.[8] Additionally, defining relevant factors contributing to HIV replication within the gut could identify new targets for the development of novel non-vaccine biomedical interventions for HIV prevention and treatment and may result in data highly relevant to the establishment of gut HIV reservoirs.
Longitudinal cohorts following HIV negative individuals at high risk of HIV acquisition have provided profound insight into the earliest systemic immune events post-HIV infection, relative to pre-exposure time-points.[9, 10] However, similar analyses of gut tissue in human cohorts have not been performed. As an alternative strategy to identify signatures within the rectal mucosa associated with support or restriction of HIV replication, we utilized the ex vivo human rectal explant model of HIV infection. This system was developed to interrogate the earliest immune responses within rectal mucosal tissues after HIV exposure, to evaluate novel anti-HIV therapeutics, and, within our lab, to identify immune cell subsets associated with support or suppression of HIV production.[11–17] Here, we performed bulk RNAseq transcriptomic analyses on rectal biopsies donated by HIV negative men (Fig 1A). In parallel, biopsies were challenged with HIV ex vivo. By associating these two data sets, we then identified transcriptomic features of rectal mucosal tissues capable of supporting higher levels of HIV replication prior to exposure.
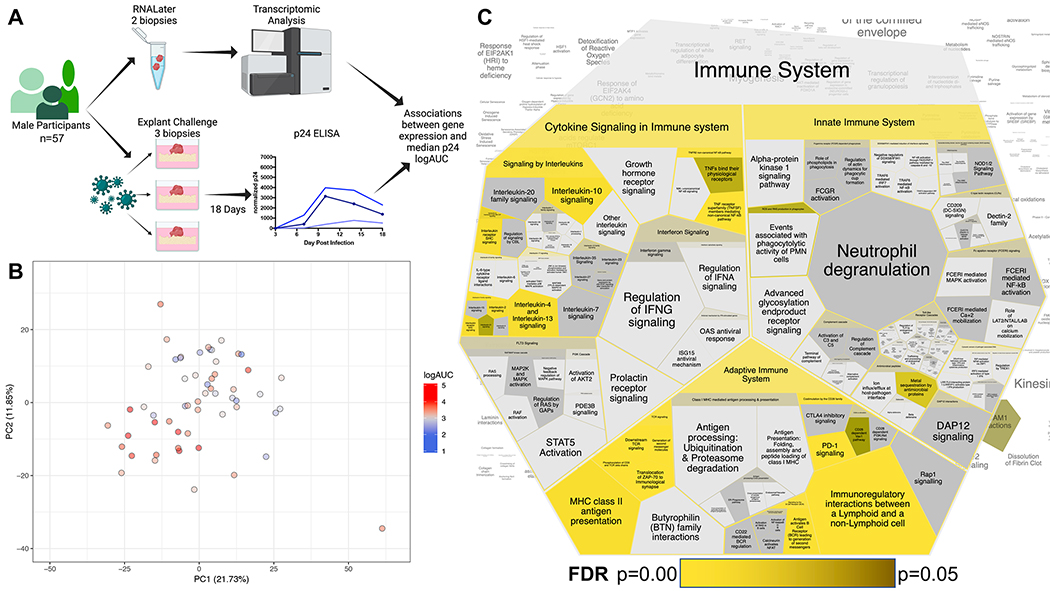
Figure 1. RNAseq transcriptomic analyses and ex vivo challenge of rectal mucosal tissue reveals associations with increased HIV replication.
A) Schematic of study procedures created with BioRender.com, in part. B) PCA plot illustrating diversity of participant rectal mucosal transcriptomes. Circle color is indicative of participant median p24 logAUC on a continuous gradient scale from low (blue) to high (red). C) Voroni diagram illustrating ‘Immune System’ pathways after Reactome analysis of the 181 significantly upregulated differentially expressed genes. Each cell represents a sub-pathway. Cell size is proportional to the proteins/molecules annotated within the pathway, not the query set. Significance of pathway enrichment is highlighted in yellow (most significant entities FDR) gradient to yellow-black (p=0.05), non-significant pathways remain gray.
Methods
This study obtained approval from Emory University Institutional Review Board (IRB) and is a subanalysis of a larger cohort study designed to better understand the unique rectal mucosal immune environment among cisgender MSM. Thus, cisgender men who did and did not engage in receptive anal intercourse (RAI) were included. Informed consent was obtained from all participants. Participants were aged 18–65 years (Fig 1A, n=57, median 40 years), from the Atlanta metropolitan area, and tested negative for rectal gonorrhea, chlamydia, and syphilis at the time of mucosal sampling. Rectal biopsies were collected via rigid sigmoidoscopy with no prior bowel preparation.
At the time of sampling, two biopsies from each participant were placed in RNALater (Invitrogen, #AM7021) and stored at −80°C. Both biopsies were homogenized in 350 μL Buffer RLT and extracted (Qiagen, RNeasy Micro kit) with on-column DNase digestion. RNA quality was assessed (Agilent, Bioanalyzer) and 10 ng of total RNA was used for cDNA synthesis (Takara Bio, Clontech SMART-Seq v4 Ultra Low Input RNA kit). Amplified cDNA was fragmented and appended with dual-indexed bar codes (Illumina, NexteraXT DNA Library Preparation kit). Libraries were validated by capillary electrophoresis (Agilent, TapeStation 4200), pooled at equimolar concentrations, and sequenced (Illumina HiSeq3000 at 100SR), yielding ~18 million reads/sample. Alignment was performed using STAR version 2.5.2b[15] and transcripts were annotated using GRCh38. Transcript abundance estimates were calculated internal to the STAR aligner using the algorithm of htseq-count,[18] GEO database: GSE211922.
For the explant challenge, three biopsies from each participant were weighed and challenged with 102.8 TCID50 HIV-1 BaL for 2 h (37°C, 5% CO2), placed on a collagen raft, and sampled longitudinally, as previously described.[17] Supernatants were collected Days 3, 7, 10, 14, 18 post-challenge, and stored at −30° C until p24 was quantified via ELISA (ABL, Inc. #5447), and normalized to biopsy weight. Log area under the curve (logAUC) was calculated for each challenge, and median logAUC from the infected replicates was used for further analyses
Neither age, reported race, RAI, CMV, HSV-1, nor HSV-2 status associated with logAUC (two-stage linear step-up method of Benjamini, Krieger and Yekutieli; False Discovery Rate = 1%; q at least 0.01. Clustering was visualized by covariance principal components analysis (PCA) using the 500 highest variance genes. Considering that race is social construct which could influence analyses in an underappreciated manner, we conservatively considered race as a covariate for differential gene expression, which was determined as a continuous covariate using DESeq2.[19] To identify the genes of greatest significance, differentially expressed genes (DEGs) were defined as i) base mean expression ≥20, ii) log2fold change ±0.58, iii) a conservative adjusted p-value ≤0.01. From the resulting list of significant DEGs (181 DEGs upregulated, 2 DEGs downregulated), the 181 upregulated DEG set was utilized as input for ReactomeFIViz app within Cytoscape Version 3.8.2.[20–22] Significant Reactome pathways were illustrated with Reacfoam. Pathways with entities false discovery rates (FDR) ≤0.01 (probability of overlap between query and pathway occurred by chance, corrected for multiple comparisons) are listed in Table 1.
Table 1.
Significant Reactome pathways with associated HitGenes.
| REACTOME PATHWAY | FDR | HITGENES |
|---|---|---|
| ADAPTIVE IMMUNE SYSTEM | 3.06E-14 | TRAC, TRBC1, HLA-DOA, HLA-DOB, LAG3, PRKCB, BTK, BLK, DAPP1, RASGRP2, CD1C, RASGRP1, CD79B, INPP5D, CD19, SLAMF6, ICOS, SH2D1A, NFATC1, PTPRC, CD28, HLA-DRA, CD22, ITK, CD40, PIK3CD, CD3G, CD3E, ITGAL, CD3D, ZAP70, LCK, EVL, WAS, FYB, HLA-DMA, HLA-DMB, KLRG1, CD74, CD247, PDCD1, CLEC2D |
| GENERATION OF SECOND MESSENGER MOLECULES | 1.53E-13 | TRAC, TRBC1, HLA-DRA, ITK, CD3G, CD3E, CD3D, ZAP70, LCK, EVL, WAS, FYB, CD247 |
| TCR SIGNALING | 7.16E-10 | TRAC, TRBC1, INPP5D, PTPRC, HLA-DRA, ITK, CD3G, CD3E, CD3D, ZAP70, LCK, EVL, WAS, FYB, CD247 |
| TRANSLOCATION OF ZAP-70 TO IMMUNOLOGICAL SYNAPSE | 1.02E-09 | TRAC, TRBC1, HLA-DRA, CD3G, CD3E, CD3D, ZAP70, LCK, CD247 |
| PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | 1.82E-09 | TRAC, TRBC1, PTPRC, HLA-DRA, CD3G, CD3E, CD3D, LCK, CD247 |
| CYTOKINE SIGNALING IN IMMUNE SYSTEM | 1.82E-09 | TNF, TNFRSF4, GBP5, EBI3, GATA3, INPP5D, TNFRSF9, LCN2, HLA-DRA, CIITA, CD40, PIK3CD, CXCL1, TNFRSF13C, JAK3, FCER2, LCK, LTA, LCP1, LTB, BIRC3, CCL4, IL21R, IL12RB1, GBP4, NOS2, CCL20, BATF, IL2RB, PTPN7 |
| PD-1 SIGNALING | 2.02E-09 | TRAC, TRBC1, HLA-DRA, CD3G, CD3E, CD3D, LCK, CD247, PDCD1 |
| COSTIMULATION BY THE CD28 FAMILY | 2.52E-08 | TRAC,T RBC1, ICOS, CD28, HLA-DRA, CD3G, CD3E, CD3D, LCK, CD247, PDCD1 |
| IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON-LYMPHOID CELL | 3.67E-07 | TRAC, TRBC1, CD1C, CD19, SLAMF6, SH2D1A, CD22, CD40, CD3G, CD3E, ITGAL, CD3D, KLRG1, CD247, CLEC2D |
| SIGNALING BY INTERLEUKINS | 8.50E-06 | TNF, EBI3, GATA3, INPP5D, LCN2, PIK3CD, CXCL1, JAK3, FCER2, LCK, LCP1, CCL4, IL21R, IL12RB1, NOS2, CCL20, BATF, IL2RB, PTPN7 |
| TNF RECEPTOR SUPERFAMILY (TNFSF) MEMBERS MEDIATING NON-CANONICAL NF-KB PATHWAY | 3.16E-05 | CD40, TNFRSF13C, LTA, LTB, BIRC3 |
| DOWNSTREAM TCR SIGNALING | 4.49E-05 | TRAC, TRBC1, INPP5D, HLA-DRA, CD3G, CD3E, CD3D, LCK, CD247 |
| INTERLEUKIN-2 FAMILY SIGNALING | 1.82E-04 | INPP5D, PIK3CD, JAK3, LCK, IL21R, IL2RB |
| TNFR2 NON-CANONICAL NF-KB PATHWAY | 2.65E-04 | TNF, TNFRSF4, TNFRSF9, CD40, TNFRSF13C, LTA, LTB, BIRC3 |
| SIGNALING BY THE B CELL RECEPTOR (BCR) | 3.48E-04 | PRKCB, BTK, BLK, DAPP1, RASGRP1, CD79B, CD19, NFATC1, CD22, PIK3CD |
| CHEMOKINE RECEPTORS BIND CHEMOKINES | 6.22E-04 | CXCL1, CXCL3, CXCR4, CXCR6, CCL4, CCL20 |
| INNATE IMMUNE SYSTEM | 8.28E-04 | C4A, TBC1D10C, BTK, HVCN1, PLD4, RASGRP2, RASGRP1, CD19, LY86, NFATC1, PTPRC, LCN2, ARHGAP9, ITK, CD3G, CXCL1, ITGAL, RAC2, CR1, AIM2, LCK, LTF, BIRC3, WAS, NLRC3, NOS2, CD247 |
| ANTIGEN ACTIVATES B CELL RECEPTOR (BCR) LEADING TO GENERATION OF SECOND MESSENGERS | 1.15E-03 | BTK, BLK, DAPP1, CD79B, CD19, CD22, PIK3CD |
| INTERLEUKIN-10 SIGNALING | 2.18E-03 | TNF, CXCL1, FCER2, CCL4, CCL20 |
| MHC CLASS II ANTIGEN PRESENTATION | 2.18E-03 | HLA-DOA, HLA-DOB, LAG3, HLA-DRA, HLA-DMA, HLA-DMB, CD74 |
| INTERLEUKIN-4 AND INTERLEUKIN-13 SIGNALING | 2.57E-03 | TNF, GATA3, LCN2, JAK3, FCER2, NOS2, BATF |
| INTERLEUKIN RECEPTOR SHC SIGNALING | 2.57E-03 | INPP5D, PIK3CD, JAK3, IL2RB |
| INTERLEUKIN-2 SIGNALING | 3.67E-03 | JAK3, LCK, IL2RB |
Results
Separation and clustering of the rectal mucosal tissue transcriptomes that supported a spectrum of lower (blue) to higher (red) p24 production in parallel biopsies in the rectal explant model (median logAUC 3.3, range 2.04–4.38) was illustrated by PCA (Fig 1B). There were 183 DEGs (181 DEGs upregulated, 2 DEGs downregulated; padj≤0.01) associated with higher p24 accumulation in the ex vivo challenge model. The bias towards upregulated DEGs suggests biological processes facilitating HIV replication, as opposed to the absence of anti-HIV biological processes within the rectal mucosal tissue. Indeed, neither of the two downregulated DEG, CHRNA1 (log2fold=−1.14, padj=0.003) and SERPINA6 (log2fold=−0.89, padj=0.007), has been associated with restricting HIV replication. Conversely, the upregulated DEG identified in this study with the greatest significance, IDO1, codes for an enzyme in the kynurenine pathway important in suppression of inflammation and the development of immune tolerance (log2fold=2.09, padj=2.55E-07)[23] and has been positively associated with HIV pathogenesis after HIV infection.[24, 25] Additional genes involved in tryptophan catabolism were also positively associated with p24 production, but did not meet our conservative threshold for DEG significance (TDO2, KMO, KYNU, padj<0.05).[26]
To determine which biological pathways were represented by the 181 upregulated DEG, these genes were used as input into ReactomeFIViz. All of the 23 Reactome pathways identified (conservative entities FDR≤0.01) implicated enrichment of immunological pathways (Fig 1C, Table 1). Because CD4+ T cells are a primary target of HIV-1 infection, the identification of significant T cell-associated pathways (e.g. ‘TCR signaling’) was expected. T cell activation genes that were associated with increased HIV replication included PDCD1/PD1 (log2fold=1.13, padj=0.003), TIGIT (log2fold=0.74, padj=0.004), CTLA4 (log2fold=1.18, padj=0.0007), TNFRSF4/OX40 (log2fold=0.58, padj=0.004), and TNFRSF9/4–1BB (log2fold=1.15, padj=0.005), as well as CD2 (log2fold=0.61, padj=0.002) and CD6 (log2fold=0.69, padj=0.005). Transcription factors often associated with specific T cell lineages also correlated with p24 production including Th1-associated TBX21/T-bet (log2fold=0.73, padj=0.002); Treg-associated FOXP3 (log2fold=0.85, padj=0.003); Th2-associated GATA3 (log2fold=0.68, padj=0.007); and CD8-associated EOMES (log2fold=1.04, padj=0.003). Th17-associated RORC (log2fold=−0.12, padj=0.82) was not significant.
Additional significant Reactome pathways included Antigen Presenting Cell (APC)/B cell pathways and cytokine/chemokine pathways (Table 1). Previous studies within our lab identified B cells within the gut, specifically CD1c+ B cells, as being associated with p24 production in the rectal explant model,[17] and CD1C was also associated with p24 here (log2fold=1.10, padj=0.003). Additional DEGs of interest included genes necessary for B cell-T cell interactions, such as CD40 (log2fold=0.72, padj=0.0004), CD83 (log2fold=0.93, padj=0.003), and IL4I1 (log2fold=0.96, padj=0.001). This suggests a gut environment primed for T cell-B cell interactions might support higher levels of p24 production, perhaps due to trans infection of CD4 T cells. Cytokine/chemokine DEG of interested included TNF (log2fold=0.74, padj=0.002), CCL4/MIP-1β (log2fold=0.66, padj=0.009), CCL20/MIP-3α (log2fold=1.49, padj=7.25E-06), CXCL1/GROα (log2fold=1.22, padj=0.001), and CXCL3/MIP-2β (log2fold=0.94, padj=0.007). While these molecules have been associated with HIV pathology,[27–30] transcriptomic analysis presented here was performed on uninfected biopsies. Thus, these DEGs may be indicative of rectal mucosal immunologic differences prior to exposure that contribute to HIV replicative capacity within this tissue compartment.
Discussion
This study, which consists of transcriptomic analysis of rectal mucosal tissue prior to prior exposure paired with the ex vivo HIV challenge model, suggests that there are immunologic features of healthy, human gut tissue that are more conducive to HIV replication. We demonstrate, for the first time, that gene signatures of T cell activation, genes critical for APC/B cell interactions with T cells, and chemokine transcription within the rectal mucosa are associated with HIV replication within this highly relevant tissue compartment. The utility of this ex vivo rectal explant challenge model is highlighted by the numerous DEGs identified here that were also identified as genes relevant during hyperacute HIV infection with single-cell transcriptomic analyses of PBMC in another study.[9] This includes genes necessary for T cell activation such as CD3D, CD3G, TRAC, CD2, FYB, and TRBC2, as well as APC/B cell-associated genes CD19, DAPP1, CD74, and HLA-DRA.[9] Our analysis also revealed associations between CCL4, CXCL1, and CXCL3, and p24 production which were also present within monocyte modules of Kazer et al.[9] Thus, it is likely that the pre-existing immunological environment within the gut sets the stage for subsequent rectal mucosal tissue responses to an HIV exposure as we demonstrate here, and this has relevance for systemic HIV pathology during hyperacute infection. These early/hyperacute events, specifically within gut tissue, are critical determinants of i) the viral set-point and subsequent disease-course, and ii) the establishment of the HIV reservoir, a substantial barrier to HIV cure strategies.[31–34] Of note, while all participants in our study were men, the participants in the Kazer study were women.[9] These potential differences underscore the need to include diverse participants in future studies to identify potential sex-, gender-, and tissue-specific DEGs associated with HIV replication to elucidate appropriate HIV prevention and treatment approaches for specific populations.[35]
These data also support further utilization of the rectal explant model in future mechanistic studies. Previous work performed by our group utilized flow cytometry to identify a significant association between an innate-like population of B cells within the rectal mucosa and p24 production within the explant model.[17] Here, we again identified APC/B cell pathways enriched in biopsies that supported higher levels of p24 production, suggesting B cells might contribute to HIV replication within the rectal mucosa in an underappreciated manner. As hypothesized previously and supported further here, CD1c+ B cells within the three-dimensional structure of the gut (and perhaps other relevant tissues such as the female reproductive tract) could bind to HIV particles, and facilitate HIV replication via trans infection of CD4+ T cells.[17, 36] This could be highly relevant to HIV prevention efforts, as trans infection is a highly potent means of propagating HIV infection,[37] and reduces the neutralization capacity of some broadly neutralizing antibodies.[38]
In conclusion, the human rectal explant model of HIV infection has provided a unique opportunity to elucidate transcriptomic signatures within the rectal mucosa of healthy men whose gut tissues supported higher levels of HIV replication. Studies are ongoing within our laboratory to explore these findings in further detail. We also seek to determine which factors could influence this observed spectrum of p24 production, such as age, hormones, and microbiome composition. Future studies can then interrogate identified pathways or genes of interest and formulate biomedical interventions to prevent rectal HIV transmission and establishment of tissue reservoirs.
Acknowledgements
P.K.A., C.F.K.: conceptualization; S.A.S., P.K.A., C.F.K.: methodology; S.A.S., C.F.K.: original draft; S.A.S., G.K.T., C.F.K.: formal analysis; C.F.K.: funding acquisition, project administration; S.A.S., P.M.M., CMG: investigation; S.A.S., P.M.M., P.K.A., C.G.A., G.K.T., S.E.B., R.R.A., C.F.K.: manuscript review & editing.
This work was funded by NIH R01 AI108335 and HD092033 (CFK). Next generation sequencing services were provided by the Emory National Primate Research Center Genomics Core, which is supported in part by NIH P51 OD011132. CMG was supported by T32 DK108735 and K12 HD085850. Additional services and support made available by Emory CFAR (P30 AI050409).
Funding Sources:
This work was funded by NIH R01 AI108335 and HD092033 (CFK), T32 DK108735 (CGA), K12 HD085850 (CGA), NIH P51 OD011132, and P30 AI050409.
References
- 1.Patel P, Borkowf CB, Brooks JT, Lasry A, Lansky A, Mermin J. Estimating per-act HIV transmission risk: a systematic review. Aids 2014; 28(10):1509–1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.CDC. Diagnoses of HIV infection in the United States and dependent areas, 2018. HIV Surveillance Report 2019. [Google Scholar]
- 3.Kalichman SC, Simbayi LC, Cain D, Jooste S. Heterosexual anal intercourse among community and clinical settings in Cape Town, South Africa. Sex Transm Infect 2009; 85(6):411–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gorbach PM, Manhart LE, Hess KL, Stoner BP, Martin DH, Holmes KK. Anal intercourse among young heterosexuals in three sexually transmitted disease clinics in the United States. Sex Transm Dis 2009; 36(4):193–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hess KL, DiNenno E, Sionean C, Ivy W, Paz-Bailey G, Group NS. Prevalence and Correlates of Heterosexual Anal Intercourse Among Men and Women, 20 U.S. Cities. AIDS Behav 2016; 20(12):2966–2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Owen BN, Elmes J, Silhol R, Dang Q, McGowan I, Shacklett B, et al. How common and frequent is heterosexual anal intercourse among South Africans? A systematic review and meta-analysis. J Int AIDS Soc 2017; 19(1):21162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sugaya M, Lore K, Koup RA, Douek DC, Blauvelt A. HIV-infected Langerhans cells preferentially transmit virus to proliferating autologous CD4+ memory T cells located within Langerhans cell-T cell clusters. J Immunol 2004; 172(4):2219–2224. [DOI] [PubMed] [Google Scholar]
- 8.Chamcha V, Reddy PBJ, Kannanganat S, Wilkins C, Gangadhara S, Velu V, et al. Strong TH1-biased CD4 T cell responses are associated with diminished SIV vaccine efficacy. Sci Transl Med 2019; 11(519). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kazer SW, Aicher TP, Muema DM, Carroll SL, Ordovas-Montanes J, Miao VN, et al. Integrated single-cell analysis of multicellular immune dynamics during hyperacute HIV-1 infection. Nat Med 2020; 26(4):511–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hassan AS, Hare J, Gounder K, Nazziwa J, Karlson S, Olsson L, et al. A Stronger Innate Immune Response During Hyperacute Human Immunodeficiency Virus Type 1 (HIV-1) Infection Is Associated With Acute Retroviral Syndrome. Clin Infect Dis 2021; 73(5):832–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abner SR, Guenthner PC, Guarner J, Hancock KA, Cummins JE, Jr., Fink A, et al. A human colorectal explant culture to evaluate topical microbicides for the prevention of HIV infection. J Infect Dis 2005; 192(9):1545–1556. [DOI] [PubMed] [Google Scholar]
- 12.Elliott J, Fulcher JA, Ibarrondo FJ, Tanner K, McGowan I, Anton PA. Comparative Assessment of Small and Large Intestine Biopsies for Ex Vivo HIV-1 Pathogenesis Studies. AIDS Res Hum Retroviruses 2018; 34(10):900–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anton PA, Cranston RD, Kashuba A, Hendrix CW, Bumpus NN, Richardson-Harman N, et al. RMP-02/MTN-006: A phase 1 rectal safety, acceptability, pharmacokinetic, and pharmacodynamic study of tenofovir 1% gel compared with oral tenofovir disoproxil fumarate. AIDS Res Hum Retroviruses 2012; 28(11):1412–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dezzutti CS, Yandura S, Wang L, Moncla B, Teeple EA, Devlin B, et al. Pharmacodynamic Activity of Dapivirine and Maraviroc Single Entity and Combination Topical Gels for HIV-1 Prevention. Pharm Res 2015; 32(11):3768–3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dezzutti CS, Russo J, Wang L, Abebe KZ, Li J, Friend DR, et al. Development of HIV-1 rectal-specific microbicides and colonic tissue evaluation. PLoS One 2014; 9(7):e102585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheeseman HM, Olejniczak NJ, Rogers PM, Evans AB, King DFL, Ziprin P, et al. Broadly Neutralizing Antibodies Display Potential for Prevention of HIV-1 Infection of Mucosal Tissue Superior to That of Nonneutralizing Antibodies. J Virol 2017; 91(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith SA, Murray PM, Amancha PK, Ackerley CG, Hu YJ, Amara RR, et al. Ex vivo rectal explant model reveals potential opposing roles of Natural Killer cells and Marginal Zone-like B cells in HIV-1 infection. Sci Rep 2020; 10(1):20154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 2015; 31(2):166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014; 15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haw R, Loney F, Ong E, He Y, Wu G. Perform Pathway Enrichment Analysis Using ReactomeFIViz. Methods Mol Biol 2020; 2074:165–179. [DOI] [PubMed] [Google Scholar]
- 21.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003; 13(11):2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jassal B, Matthews L, Viteri G, Gong C, Lorente P, Fabregat A, et al. The reactome pathway knowledgebase. Nucleic Acids Res 2020; 48(D1):D498–D503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Favre D, Mold J, Hunt PW, Kanwar B, Loke P, Seu L, et al. Tryptophan catabolism by indoleamine 2,3-dioxygenase 1 alters the balance of TH17 to regulatory T cells in HIV disease. Sci Transl Med 2010; 2(32):32ra36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmidt SV, Schultze JL. New Insights into IDO Biology in Bacterial and Viral Infections. Front Immunol 2014; 5:384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baer SL, Colombo RE, Johnson MH, Wakade S, Pacholczyk G, Newman-Whitlow C, et al. Indoleamine 2,3 dioxygenase, age, and immune activation in people living with HIV. J Investig Med 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ye Z, Yue L, Shi J, Shao M, Wu T. Role of IDO and TDO in Cancers and Related Diseases and the Therapeutic Implications. J Cancer 2019; 10(12):2771–2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pasquereau S, Kumar A, Herbein G. Targeting TNF and TNF Receptor Pathway in HIV-1 Infection: from Immune Activation to Viral Reservoirs. Viruses 2017; 9(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cocchi F, DeVico AL, Garzino-Demo A, Arya SK, Gallo RC, Lusso P. Identification of RANTES, MIP-1 alpha, and MIP-1 beta as the major HIV-suppressive factors produced by CD8+ T cells. Science 1995; 270(5243):1811–1815. [DOI] [PubMed] [Google Scholar]
- 29.Wang Z, Shang H, Jiang Y. Chemokines and Chemokine Receptors: Accomplices for Human Immunodeficiency Virus Infection and Latency. Front Immunol 2017; 8:1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lane BR, Strieter RM, Coffey MJ, Markovitz DM. Human immunodeficiency virus type 1 (HIV-1)-induced GRO-alpha production stimulates HIV-1 replication in macrophages and T lymphocytes. J Virol 2001; 75(13):5812–5822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lavreys L, Baeten JM, Chohan V, McClelland RS, Hassan WM, Richardson BA, et al. Higher set point plasma viral load and more-severe acute HIV type 1 (HIV-1) illness predict mortality among high-risk HIV-1-infected African women. Clin Infect Dis 2006; 42(9):1333–1339. [DOI] [PubMed] [Google Scholar]
- 32.Chun TW, Engel D, Berrey MM, Shea T, Corey L, Fauci AS. Early establishment of a pool of latently infected, resting CD4(+) T cells during primary HIV-1 infection. Proc Natl Acad Sci U S A 1998; 95(15):8869–8873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McMichael AJ, Borrow P, Tomaras GD, Goonetilleke N, Haynes BF. The immune response during acute HIV-1 infection: clues for vaccine development. Nat Rev Immunol 2010; 10(1):11–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ananworanich J, Schuetz A, Vandergeeten C, Sereti I, de Souza M, Rerknimitr R, et al. Impact of multi-targeted antiretroviral treatment on gut T cell depletion and HIV reservoir seeding during acute HIV infection. PLoS One 2012; 7(3):e33948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sekabira R, McGowan I, Yuhas K, Brand RM, Marzinke MA, Manabe YC, et al. Higher colorectal tissue HIV infectivity in cisgender women compared with MSM before and during oral preexposure prophylaxis. AIDS 2021; 35(10):1585–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fourcade L, Sabourin-Poirier C, Perraud V, Faucher MC, Chagnon-Choquet J, Labbe AC, et al. Natural Immunity to HIV is associated with Low BLyS/BAFF levels and low frequencies of innate marginal zone like CD1c+ B-cells in the genital tract. PLoS Pathog 2019; 15(6):e1007840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gerberick A, DeLucia DC, Piazza P, Alaoui-El-Azher M, Rinaldo CR, Sluis-Cremer N, et al. B Lymphocytes, but Not Dendritic Cells, Efficiently HIV-1 Trans Infect Naive CD4(+) T Cells: Implications for the Viral Reservoir. mBio 2021; 12(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dufloo J, Bruel T, Schwartz O. HIV-1 cell-to-cell transmission and broadly neutralizing antibodies. Retrovirology 2018; 15(1):51. [DOI] [PMC free article] [PubMed] [Google Scholar]