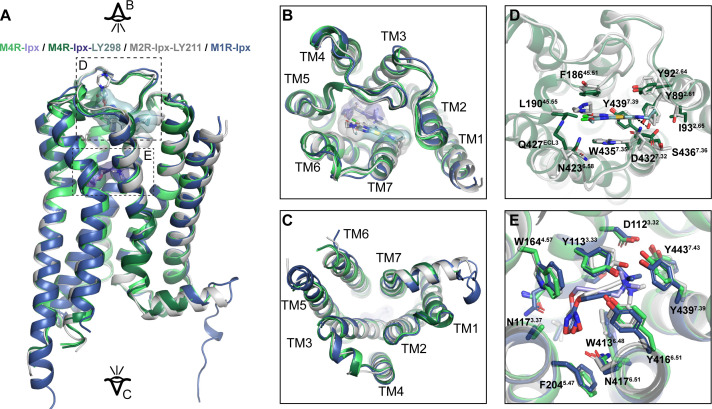
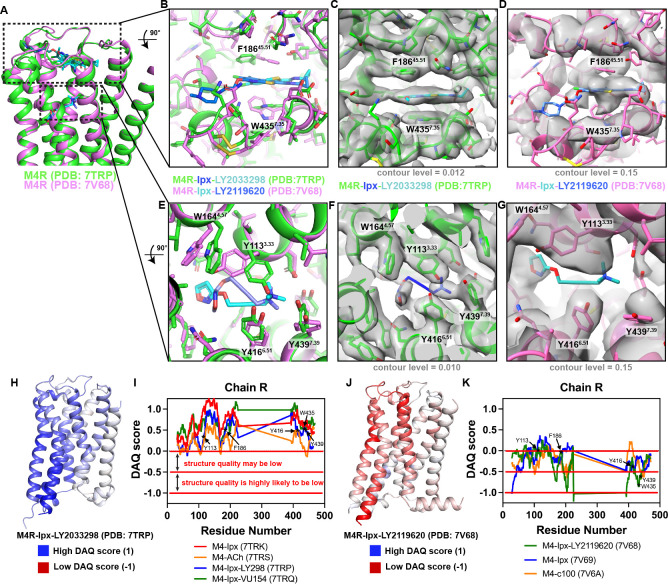
(
A) Comparison of LY298-Ipx bound M
4 mAChR structure (PDB: 7TRP, receptor colored green, Ipx blue, and LY298 cyan) to the LY2119620-Ipx bound M
4 mAChR structure (PDB: 7V68, receptor colored pink, Ipx cyan, and LY2119620 blue) (
Wang et al., 2022). (
B–D) View of the allosteric binding site from the top of the receptor. (
B) Comparison of key allosteric residues F186
45.51 and W435
7.35 showing different positions of the residues between M
4 mAChR structures. (
C) Overlay of the EM map (EMD-26100, colored gray) onto the LY298-Ipx bound M
4 mAChR structure contoured at 0.012. (
D) Overlay of the EM map (EMD-31738, colored gray) onto the LY2119620-Ipx bound M
4 mAChR structure contoured at 0.15. There is a lack of EM density surrounding the allosteric residues F186
45.51 and W435
7.35 at this level of contour and all others. (
E–G) View of the orthosteric binding site from the top of the receptor. (
E) Comparison of key orthosteric binding site residues. (
F) Related to (
C) with view from orthosteric site and the EM-map contoured at 0.010. (
G) Related to (
D) with view from the orthosteric site with mismodeled residues. (
H–K) DAQ scores provide an estimation of the local quality of protein models from cryo-electron microscopy (cryo-EM) maps on a per residue basis. DAQ scores were determined from the DAQ web server using the recommended default settings (
Terashi et al., 2022). (
H, J) DAQ scores from the analysis of (
H) the LY298-Ipx-M
4R-G
i1 complex and (
J) the LY2119620-Ipx-M
4R-G
i1 complex mapped onto the cartoon of the receptor chain and color coded by score. A DAQ score that is positive (colored blue at values of 1) indicates a correct assignment. A DAQ score near 0 (colored white) indicates a position in the map that lacks a distinct density pattern for the assigned amino acid. DAQ scores less than 0 (colored red at –1) indicate a position that could be misassigned or poorly fit. (
I) DAQ scores for all four M
4 mAChR structures reported in this article with DAQ scores of each Cα atom plotted for each residue. Key orthosteric and allosteric residues are denoted by asterisks. Nearly every residue has a value above 0. (
K) Similar to (
I), but for all three M
4 mAChR structures reported in
Wang et al., 2022. Very few residues have a score above 0, indicating potential issues with the model and maps.