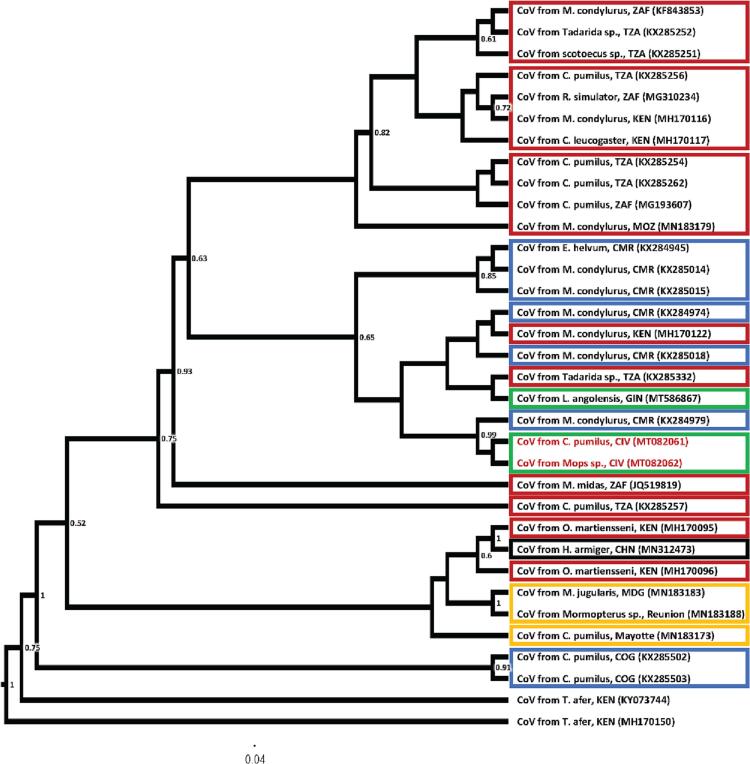
Fig. 2.
Maximum likelihood phylogenetic tree of BtKY22-like CoVs based on the amplified PCR fragment of the RdRp gene, including sequences closest (>90% identities) to Cote d'Ivoire CoV isolate from Chaerephon cf. pumilus and Mops sp.. Isolates are identified by GenBank ID and host (bat species) from which the CoV sequence was obtained as well as country three letter code (ISO 3166-1 alpha-3) or place were sampling took place are indicated. Geographic regions are indicated with boxes for sequences >90% identical with Cote d'Ivoire isolates, with green indicating West Africa, blue Central Africa, red West & South Africa, orange Islands in the Indian Ocean, and black “other continent” (Compare Supplement 5). Numbers at nodes indicate bootstrap support. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)