Figure 1.
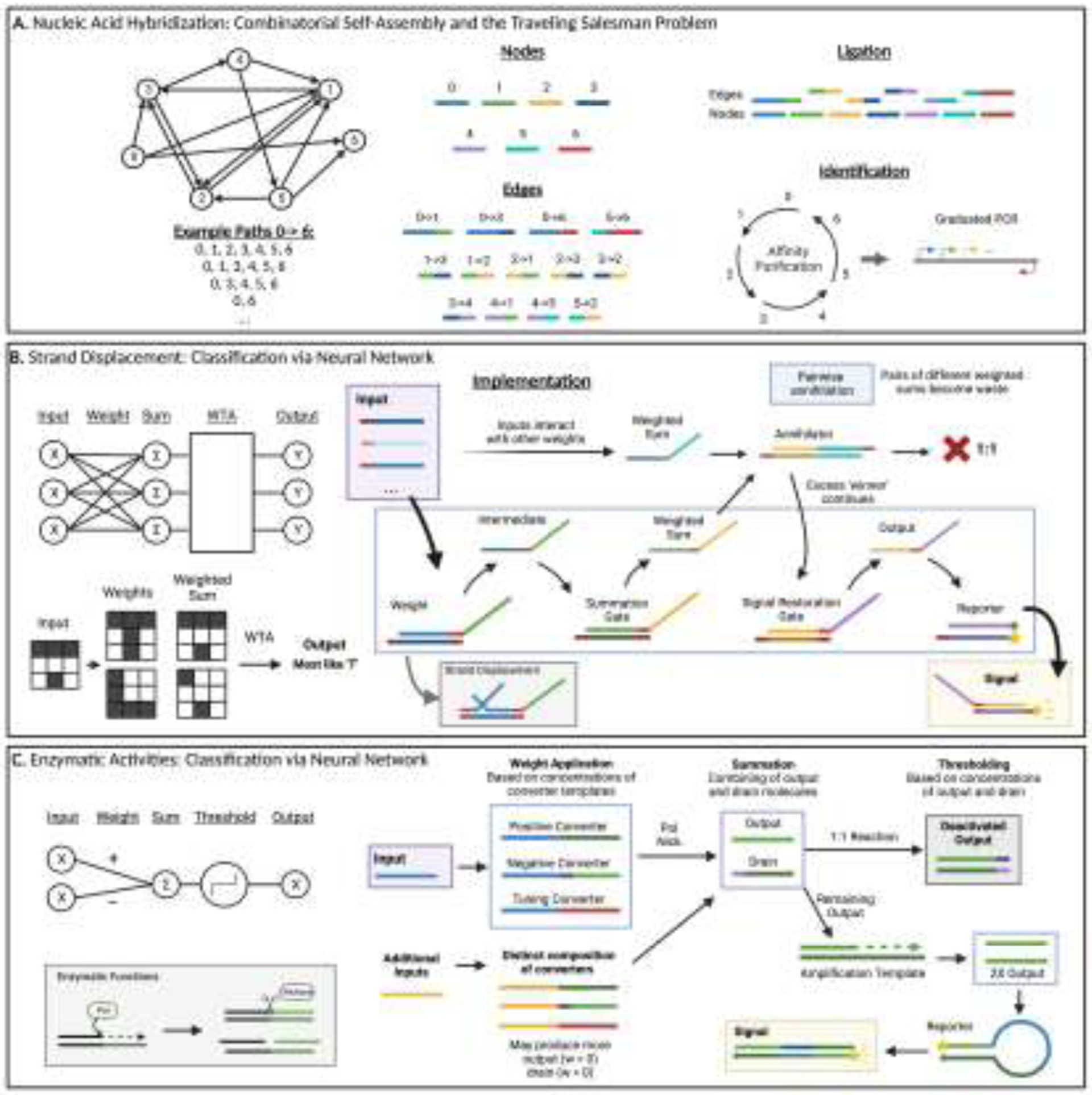
A. Nucleic Acid Hybridization: Combinatorial Self-Assembly and the Traveling Salesman Problem
Nodes were represented by a specified sequence with edge sequences complementary to both the 3’ end of its origin node sequence and the 5’ end of its destination node. Edges leading to or from 0 or 6 were complementary to the full sequence of those nodes. When combined in a one-pot ligation reaction, these complementary regions ligated to form strands encoding possible paths. The double-stranded sequences were then PCR-amplified with primers annealing to the sequences of nodes 0 and 6, , satisfying the requirement that each path enter at node 0 and exit at node 6. These products were selected for the correct size by gel electrophoresis. To isolate paths containing every node, Adelman then used sequential affinity purification to pull down each node in turn and verified the correct final sequence through graduated PCR.
B. Strand Displacement: Classification via Neural Network
Cascading strand displacement converts an input molecule into an intermediate strand through a weight molecule, in proportion to the initial concentration of the input. The intermediate interacts with a summation gate to produce a strand representing the weighted sum. This weighted sum strand participates in a pairwise annihilation reaction that performs the winner-take-all (WTA) function, in which weighted sums are compared to known digits, or ‘memories,’ and only the most similar remembered digit given as output. This occurs on the molecular level via 1:1 deactivation of distinct weighted sums through hybridization, leaving only the most prevalent weighted sum molecule active. This ‘winner’ may continue to signal restoration that produces an output strand and leads to fluorescence of a reporter molecule. Weight concentrations were assigned based on the color value (darkness of writing) of each of 100 pixels within the image. Values of input symbols were compared to the values of representative images of digits through the network, and the most similar digit reported as output.
C. Enzymatic Activities: Classification via Neural Network
The PEN DNA toolbox mechanism is used to generate a short output strand upon addition of the input strands, implementing positive weight, while also producing a drain molecule that hybridizes and deactivates the output, acting as the negative weight. These weights were tuned using additional template strands that competed for the inputs but produced a nonfunctional molecule. Interaction of the output strand and drain strand performed the thresholding, as output molecules that overcome the drain mechanism remain available for an amplification step using an additional template strand. This generates more output strands to interact with the reporter molecule to generate fluorescent signal. Created with Biorender.com
