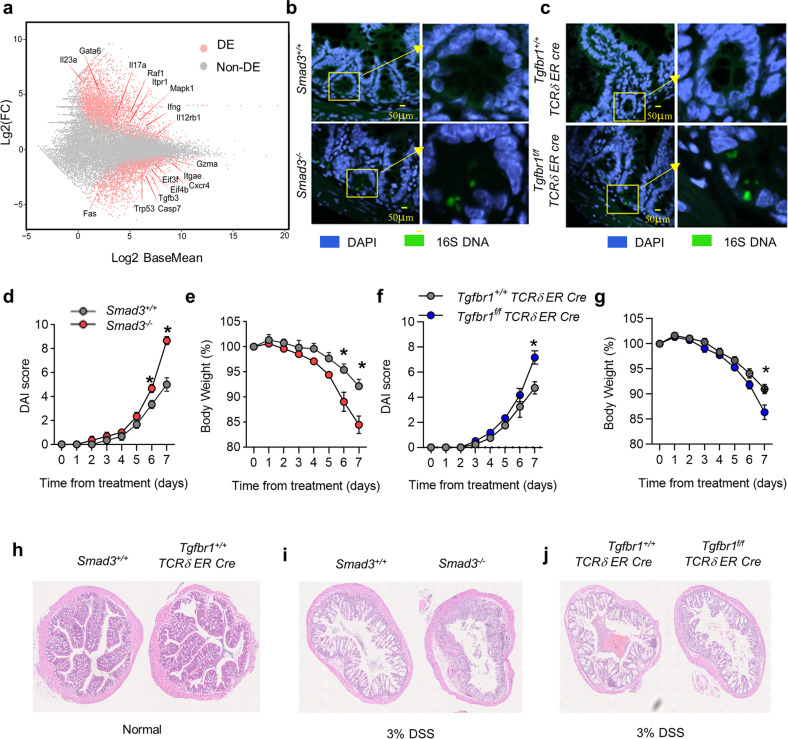
Fig. 7. Lack of TGF-β signaling in γδ IELs exacerbates DSS-induced colitis.
a The volcano plot shows the differential expression profile of genes in TCRγδ+CD8αα+ IELs from R1 WT and R1 KO mice and examined by RNA-seq. The gray dots represent non-differentially expressed genes between the two groups, while the upregulated genes in R1 KO mice are pink dots towards the upper end (with log2 fold change value above 0 on the y-axis), and the downregulated genes in R1 KO mice are pink dots towards the lower end (with log2 fold change value below 0 on the y-axis). b FISH visualizes the location of bacteria around the intestinal epithelia in the small intestine of Smad3−/− mice and age-matched littermate controls (Smad3+/+). c FISH visualizes the location of bacteria in the small intestine from 5-day-tamoxifen-treated Tgfbr1f/f TCRδ ER Cre and age-matched Tgfbr1+/+ TCRδ ER Cre littermates. d, e DAI (d) and loss of body weight (e) of Smad3−/− and Smad3+/+ mice treated with 3% DSS drinking water for 7 days. f, g DAI (f) and loss of body weight (g) of Tgfbr1f/f TCRδ ER Cre and Tgfbr1+/+ TCRδ ER Cre control mice treated with DSS for 7 days. h–j Hematoxylin and eosin (H&E) staining for colon from normal Smad3−/− and 5-day-tamoxifen-treated Tgfbr1+/+f TCRδ ER Cre mice (h), and from 3% DSS-treated Smad3−/− vs Smad3+/+ mice in d and e (i), or 3% DSS-treated Tgfbr1f/f TCRδ ER Cre versus Tgfbr1+/+ TCRδ ER Cre mice in f and g (j). *P < 0.05 (unpaired two-tailed Student’s t-test or DEseq2 statistical test). Data were representative of at least two independent experiments (means ± SEM). RNA-seq samples were collected from three or four independent experiments.