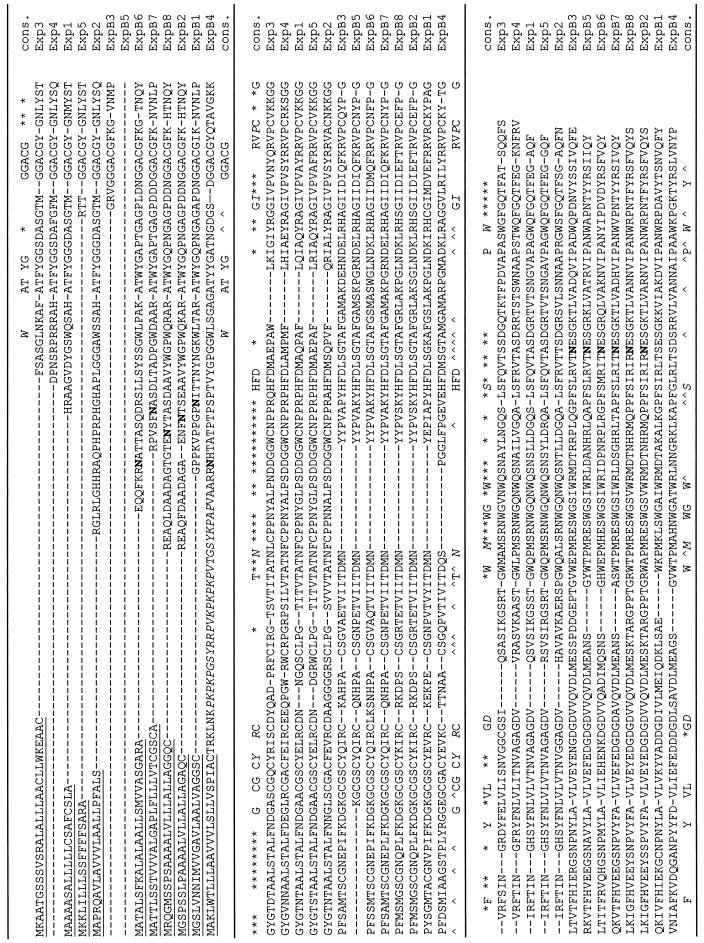
Figure 1.
Alignment of amino acid sequences predicted from maize expansin cDNAs. Sequences were aligned by MEGALIGN program (DNAstar software) using the Clustal method with the PAM250 residue weight table and default parameters. The underlined sequences represent the signal peptides predicated by the SignalP program (www.cbs.dtu.dk/services/SignalP/). Potential N-linked glycosylation sites are shown in bold and were predicted using Prosite. Symbols in the consensus sequence, Capitalized letters indicate conserved amino acids for α- and β-expansins; italicized capital letters denote conserved amino acids for α- and β-expansins with occasional variance; an asterisk denotes amino acids conserved among α-expansins; and ∧ denotes amino acids conserved among β-expansins.