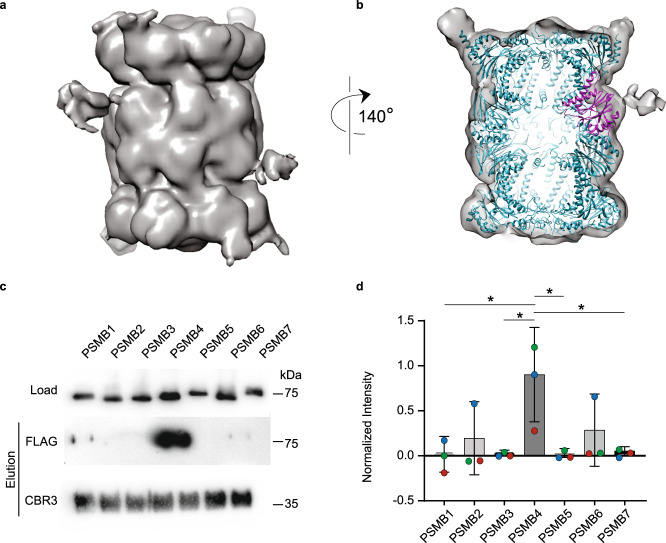
Fig. 7. CBR3 binds the 20S proteasome β-ring and specifically to the PSMB4 subunit.
a Cryo-EM structure of the rat 20S proteasome/CBR3 complex. A prominent extra density is displayed at the β-ring region. b Atomic model of the 20S proteasome structure (PDB:6TU3) (cyan) was fitted into the electron density map. The extra electron density near the PSMB4 subunit (magenta) reveals the binding site of CBR3. c Escherichia coli cell lysates overexpressing FLAG-tagged human β-subunits PSMB1 to PSMB7 were incubated with His6-CBR3 bound to Ni-beads for pull-down experiments. Bound subunits were detected using an anti-FLAG antibody. d Densitometry quantification indicated that the pull-down of PSMB4 greatly exceeds the levels of the other β-subunits. The bar graph shows the average values of each PSMB subunit intensities in the pull-down, normalized to the corresponding lysate, from three independent replicates; each replica is color-coded (red, green, and blue). Error bars represent SEM. Significance was calculated using one-way ANOVA (p-value = 0.0309), with Dunnett’s post-hoc test (* represents p-value = 0.0145, 0.0168, 0.0163, 0.0191 for the significances between PSMB4 and PSMB1, PSMB3, PSMB5, and PSMB7, respectively). Source data are provided with this paper.