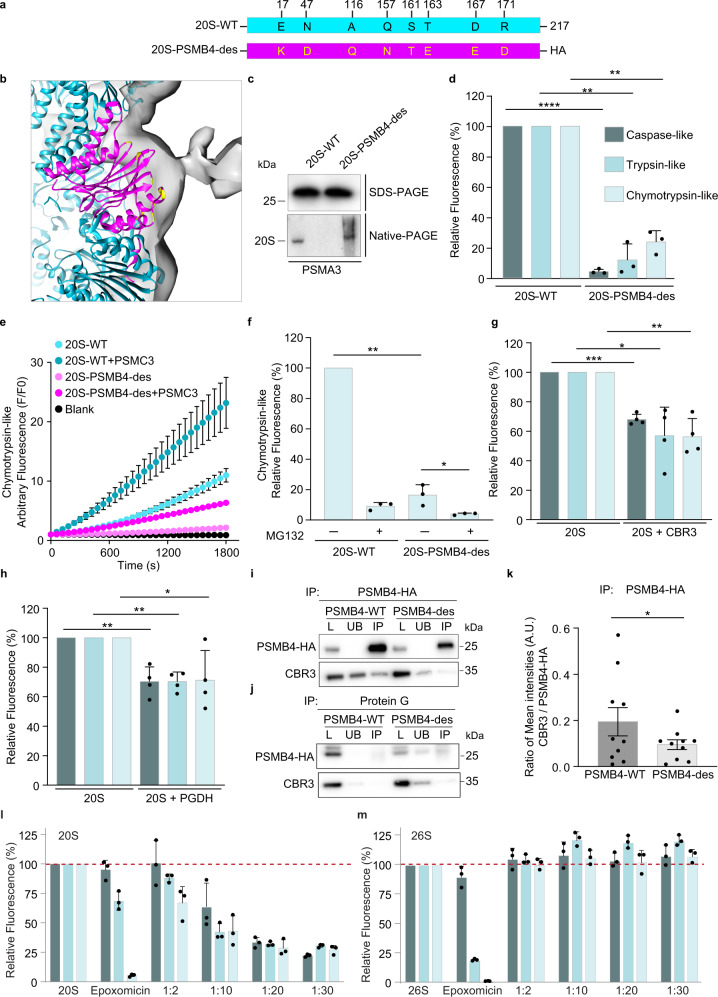
Fig. 8. CBR3 binds the PSMB4 subunit of the 20S proteasome.
a Schematic representation of human PSMB4-WT and 20S-PSMB4-des engineered to attenuate CBR3 binding. The mutated residues are highlighted in yellow. b Zoom-in view of the PSMB4 subunit (magenta) with mutated residues highlighted in yellow. c Purified proteasomes were analyzed by SDS- and native-PAGE and blotted with an anti-PSMA3 antibody, indicating the reconstitution of holo-20S proteasomes consisting of the HA-tagged PSMB4-WT or des subunit. d The chymotrypsin-, trypsin- and caspase-like activities of the purified WT 20S proteasome and those incorporating the PSMB4-des subunit were measured using fluorogenic peptide substrates. A pronounced reduction in all three enzymatic activities was detected for the 20S-PSMB4-des complex. Averaged quantification of three independent experiments; error bars represent SD. e The chymotrypsin-like activity of WT 20S proteasome and the 20S-PSMB4-des was measured in real-time in the presence and absence of the gate-opening PSMC3 C-terminus peptide. Averaged quantification of three independent experiments; error bars represent S.D. f The influence of the proteasome inhibitor, MG132, on WT 20S proteasome and on 20S-PSMB4-des was measured by using the fluorogenic peptide substrate suc-LLVY-AMC. The graph represents averaged values from three independent repeats; error bars represent SD. The impact of the CCRs g CBR3 and h PGDH on the chymotrypsin-, trypsin-, and caspase-like activities of the purified rat 20S proteasome was monitored using fluorogenic peptide substrates. Similar to the 20S-PSMB4-des complex, CBR3 and PGDH significantly reduced all three enzymatic activities of the proteasome. Bars and scatter plots represent mean values from three or four independent experiments (in f and g, h, respectively), and error bars represent SD. Measurements were subjected to two-tailed (d) and one-tailed (f–h) Student’s t-test analysis (d **** represents p-value = 0.00007, ** for trypsin-like represents p-value = 0.0047, ** for chymotrypsin-like represents p-value = 0.0031). (f ** represents p-value = 0.0011, * represents p-value = 0.0433). (g *** represents p-value = 0.0002, * represents p-value = 0.0108, ** represents p-value = 0.0027). (h ** for caspase-like represents p-value = 0.0048, ** for trypsin-like represents p-value = 0.0015, * represents p-value = 0.0326). i, j PSMB4-WT-HA or PSMB-des-HA and CBR3 were overexpressed in HEK293T cell. Lysates were subjected to IP using either anti-HA agarose (i) or uncoupled protein G beads as a control (j). Total starting lysate (L), unbound proteins (UB), and IP samples were analyzed by Western blot using anti-HA or anti-CBR3 antibodies. k Bands corresponding to the pull-down of CBR3, WT-, and des- PSMB4-HA were quantified, and the ratio of CBR3 to PSMB4-HA (WT and des) are presented as bar graph. Mean values from 10 independent experiments were subjected to paired one-tailed Student t-test analysis, * represents p-value = 0.05. Error bars represent SEM. The catalytic activities of the human l 20S and m 26S proteasomes were measured in the presence of increasing concentrations of the CCR PGDH. A concentration-dependent increase in inhibition of the 20S proteasome is detected, while the 26S proteasome is not inhibited at any concentration of PGDH tested. Bars represent mean values from three independent experiments. Error bars represent SD. Source data are provided with this paper.