Abstract
The use of an effective antimalarial drug is the cornerstone of malaria control. However, the development and spread of resistant Plasmodium falciparum strains have placed the global eradication of malaria in serious jeopardy. Molecular marker analysis constitutes the hallmark of the monitoring of Plasmodium drug-resistance. This study included 96 P. falciparum PCR-positive samples from southern Somalia. The P. falciparum chloroquine resistance transporter gene had high frequencies of K76T, A220S, Q271E, N326S, and R371I point mutations. The N86Y and Y184F mutant alleles of the P. falciparum multidrug resistance 1 gene were present in 84.7 and 62.4% of the isolates, respectively. No mutation was found in the P. falciparum Kelch-13 gene. This study revealed that chloroquine resistance markers are present at high frequencies, while the parasite remains sensitive to artemisinin (ART). The continuous monitoring of ART-resistant markers and in vitro susceptibility testing are strongly recommended to track resistant strains in real time.
Keywords: Plasmodium falciparum, drug-resistance, molecular marker, Southern Somalia
Malaria is a significant health problem in most tropical countries, including Somalia. Plasmodium falciparum is the most prevalent malaria species in sub-Saharan Africa [1]. The emergence of multidrug-resistant P. falciparum strains poses a challenge to malaria control plans and has considerably increased drug costs. This situation endangers affordable, safe, and cheap drugs in the least-developed countries wherein malaria control is ineffective [2]. The World Health Organization recommends using artemisinin (ART)-based combination therapies as the first-line treatment for uncomplicated falciparum malaria to confront the spread of treatment-resistant malaria strains [3]. In 2006, Somalia discontinued chloroquine (CQ) as the first-line therapy for uncomplicated falciparum malaria because CQ has high treatment failure rates that range from 76.5 to 88.0% in Janale, Jamame, and Jowhar. The combination of artesunate and sulphadoxine–pyrimethamine was introduced into the national malaria control program [4]. The country’s first-line treatment was later switched to artemether–lumefantrine (AL) due to numerous mutations in antifolate resistance markers [5]. ART-resistance has been reported to be strongly associated with nonsynonymous single nucleotide polymorphisms within the propeller region of the P. falciparum Kelch-13 (pfk13) gene [6]. Of the 10 validated ART-resistance mutations, 6 (M476I, P553L, R561H, P574L, C580Y, and A675V) have been found at low frequencies in 4 African countries. However, Rwanda is the only country to report delayed parasite clearance (3+ days) [7]. The emergence of pfk13 mutations is an early warning sign for Africa given that an increase in the frequency and spread of these mutations across the continent could have catastrophic consequences for malaria control. The purpose of the present study is to determine the prevalence rates of drug-resistance molecular markers (polymorphisms in the P. falciparum chloroquine resistance transporter [pfcrt] gene, P. falciparum multidrug resistance 1 [pfmdr1], P. falciparum ATPase 6 [pfatp6], and pfk13) and the copy number variation (CNV) of pfmdr1 in isolates from Afgoi and Balad in southern Somalia.
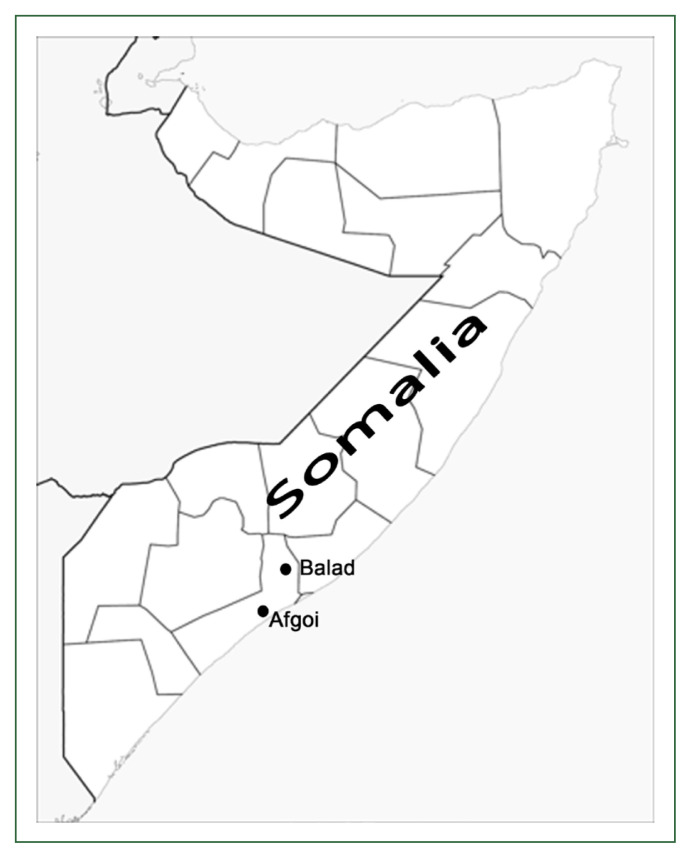
Approval for the study protocol was given by the Ethics Review Committee for Research in Human Subjects, Ministry of Health of Somalia (No. 0776/May/2018). Informed consent for study participation was obtained from all participants. A total of 150 dried blood spot samples were collected in 2018 from patients with P. falciparum infection attending malaria clinics in southern Somalia (Afgoi and Balad; Fig. 1). DNA was extracted from blood samples spotted on filter paper by using a QIAamp DNA Mini kit (Qiagen, Hilden, Germany) in accordance with the manufacturer’s protocol then used as a template for PCR amplification. The polymorphisms of pfcrt, pfmdr1, and pfatp6 were examined by using PCR with restriction fragment length polymorphism [8,9]. CNVs were assessed by utilizing a Bio-Rad iCycler (Bio-Rad, Hercules, CA, USA) [10]. The pfk13 gene was amplified by nested PCR as described by Ariey et al. [6]. Amplified products were purified by applying a QIAquick PCR purification kit (Qiagen) then sequenced by a commercial vendor (Bioneer, Daejeon, Republic of Korea). DNA sequencing was performed in 2 directions for complete coverage. The sequences of each isolate were aligned with the 3D7 reference strain (GenBank accession number, XM_001350122) by employing BioEdit software ver7.0.5.2 (Ibis Therapeutics, Carlsbad, CA, USA). Chi-square or Fisher’s exact test was used to compare the proportions of the mutated points in the 2 study sites (Afgoi vs Balad). The level of statistical significance was set at α=0.05.
Fig. 1.
Map of Somalia showing the study site.
A total of 150 microscopically confirmed P. falciparum samples were collected for the study. PCR confirmed the presence of malaria species in 67.3% of the samples (96 P. falciparum and 5 Plasmodium vivax), whereas the remaining samples were PCR-negative. Thus, 96 P. falciparum samples were included in molecular analysis. Most of the samples (81.0%) included in the molecular analysis were from the Afgoi district in the Lower Shabelle region. Eighty-six samples were successfully genotyped at all codons of pfcrt. All isolates from Balad showed mutant genotypes at codons K76T, A220S, Q271E, and R371I and wild genotypes at codon I356T. The frequencies of each mutant point showed no significant difference between Afgoi and Balad isolates (P>0.05) (Table 1). The frequency of the pfmdr1 mutation among the 85 samples that were successfully amplified is shown in Table 1. Mutations in pfmdr1 were only seen at N86Y and Y184F in the isolates from the 2 study areas. Meanwhile, the 3 other positions (S1034C, N1042D, and D1246Y) were 100.0% wild-type. Four haplotype patterns of pfmdr1 were found: NYSND (2.4%), YYSND (35.3%), NFSND (12.9%), and YFSND (49.4%). The most dominant haplotype was the YFSND double-mutated haplotype, which represents the substitution of the amino acid tyrosine for codon 86 and phenylalanine for codon 184 [8]. On the other hand, the wild NYSND haplotype had the lowest prevalence. A total of 84 isolates were subjected to pfmdr1 gene amplification analysis. Only 3 isolates from Afgoi were found to carry 2 copies of pfmdr1 (Table 1). The pfatp6 gene had mutant genotypes at codons R37K, G639D, and I898I and a wild-type S769N codon. The I898I mutation was detected in isolates from the 2 study sites, whereas mutations at codons R37K and G639D were detected only in Afgoi isolates (Table 1). All 84 samples that were successfully sequenced for P. falciparum isolates carried the wild-type alleles of pfk13.
Table 1.
Distribution of pfcrt, pfmdr1 and pfatp6 in Plasmodium falciparum isolates from southern Somalia. Data is presented as the number of isolates (%)
| Gene | Codon | Amino Acid | No. of isolates (%) | ||
|---|---|---|---|---|---|
|
| |||||
| Afgoi | Balad | Total | |||
| pfcrt | 76 | K (Wild-type) | 15 (17.4) | 0 (0.0) | 15 (17.4) |
| Tb (Mutation) | 54 (62.8) | 17 (19.8) | 71 (82.6) | ||
| 220 | A (Wild-type) | 2 (2.3) | 0 (0.0) | 2 (2.3) | |
| Sb (Mutation) | 67 (77.9) | 17 (19.8) | 84 (97.7) | ||
| 271 | Q (Wild-type) | 1 (1.2) | 0 (0.0) | 1 (1.2) | |
| Eb (Mutation) | 68 (79.1) | 17 (19.8) | 85 (98.8) | ||
| 326 | N (Wild-type) | 7 (8.1) | 11 (12.8) | 18 (20.9) | |
| Sb (Mutation) | 62 (72.1) | 6 (7.0) | 68 (79.1) | ||
| 356 | I (Wild-type) | 67 (77.9) | 17 (19.8) | 84 (97.7) | |
| Tb (Mutation) | 2 (2.3) | 0 (0.0) | 2 (2.3) | ||
| 371 | R (Wild-type) | 5 (5.8) | 0 (0.0) | 5 (5.8) | |
| Ib (Mutation) | 64 (74.4) | 17 (19.8) | 81 (94.2) | ||
|
| |||||
| pfmdr1 | 86a | N (Wild-type) | 7 (8.2) | 6 (7.1) | 13 (15.3) |
| Yb (Mutation) | 62 (72.9) | 10 (11.8) | 72 (84.7) | ||
| 184 | Y (Wild-type) | 25 (29.4) | 7 (8.2) | 32 (37.6) | |
| Fb (Mutation) | 44 (51.8) | 9 (10.6) | 53 (62.4) | ||
| 1034 | S (Wild-type) | 69 (81.2) | 16 (18.8) | 85 (100.0) | |
| 1042 | N (Wild-type) | 69 (81.2) | 16 (18.8) | 85 (100.0) | |
| 1246 | D (Wild-type) | 69 (81.2) | 16 (18.8) | 85 (100.0) | |
| Copy no. | 1 copy | 65 (77.4) | 16 (19.0) | 81 (96.4) | |
| 2 copies | 3 (3.6) | 0 (0.0) | 3 (3.6) | ||
|
| |||||
| pfatp6 | 37 | R (Wild-type) | 67 (77.9) | 16 (18.6) | 83 (96.5) |
| Kb (Mutation) | 3 (3.5) | 0 (0.0) | 3 (3.5) | ||
| 639 | G (Wild-type) | 66 (76.7) | 16 (18.6) | 82 (95.3) | |
| Db (Mutation) | 4 (4.7) | 0 (0.0) | 4 (4.7) | ||
| 769 | S (Wild-type) | 70 (81.4) | 16 (18.6) | 86 (100.0) | |
| 898 | I (Wild-type) | 46 (53.5) | 11 (12.8) | 57 (66.3) | |
|
| |||||
| Ib (Mutation) | 24 (27.9) | 5 (5.8) | 29 (33.7) | ||
Statistically significant difference between Afgoi and Balad isolates (P=0.006).
Mutation codons.
pfcr, P. falciparum chloroquine resistance transporter; pfmdr1, P. falciparum multidrug resistance 1; pfatp6, P. falciparum ATPase 6.
The molecular analysis of antimalarial drug-resistance markers provides information on parasite gene mutations and/or gene copy number changes associated with drug-resistance [11]. The pfcrt and pfmdr1 genes are generally considered markers of CQ resistance (CQR) [8]. The pfmdr1 gene and the pleiotropic drug-resistance transporter pfcrt reside on the membrane of the parasite’s digestive vacuole and are believed to prevent osmotic stress in the vacuole by exporting host-derived peptides and delivering them to the cytosol, where they are degraded into amino acids to fuel parasite growth [12]. The pfcrt gene is a stronger predictor of CQR than pfmdr1. The K76T mutation is the crucial marker of CQR, whereas the other alleles of the pfcrt gene confer a lesser degree of CQR [8]. In this study, the K76T mutation was found in 82.6% of isolates. All isolates from Balad carried the K76T mutation. The study also showed high mutation rates at codons A220S, Q271E, N326S, and R371I. The accumulation of point mutations in the pfcrt gene has been associated with a high level of CQR [13]. This finding contradicts previously reported results showing the re-emergence of CQ-sensitive P. falciparum populations after the withdrawal of CQ from the national treatment policy [14]. This situation might be due to the availability of the drug in local drug stores that leads to continued drug pressure on circulating malaria parasites [15]. Of the 5 polymorphisms of the pfmdr1 gene, no mutations were detected at positions S1034C, N1042D, or D1246Y in any isolate. The wild haplotype of these 3 points is common in CQR parasites in Africa and Asia, whereas their mutant haplotypes are common in CQR parasites from South America [16]. The present study found predominant mutations of pfmdr1 N86Y and Y184F (84.7 and 62.4%, respectively). However, in accordance with study sites, only the N86Y mutation showed a significant difference in frequencies between the isolates from Afgoi and Balad (P<0.05). This discrepancy might be due to differences in sample size (Afgoi vs. Balad=69 vs. 16). The mutation profiles of N86Y and Y184F showed similar patterns, as reported by a previous study from Tanzania [17]. The N86Y mutant allele contributes to resistance to CQ and amodiaquine but increases parasite susceptibility to lumefantrine, mefloquine, and dihydroartemisinin [16]. No previously published data on pfcrt and pfmdr1 allelic variants from Somalia are available for comparison with the present study’s findings. Among the pfmdr1 haplotypes, YFSND was the most common (49.4%), followed by YYSND (35.3%). The relatively high frequency of the double mutational haplotype detected in the present study might be due to continuous selective pressure from CQ [15]. This study also showed a low prevalence rate of 2 pfmdr1 copies (3.6%) in southern Somalia. This finding is consistent with the result of a study from Kenya, which reported a prevalence of 2 pfmdr1 copies of less than 10.0% [18]. Notably, increased pfmdr1 copy number was associated with lower parasite susceptibility to mefloquine and partial susceptibility to lumefantrine [10]. Most likely, the low rate of parasites with 2 copies of pfmdr1 is due to the fact that AL has been a part of the national policy for malaria treatment since 2016 [5]. The pfk13 and pfatp6 genes are potential markers among the candidate genes for P. falciparum ART-resistance [6,9]. Jambou et al. [9] observed that the mutation at S769N in the pfatp6 gene is associated with an increased in vitro susceptibility to artemether. Although this work discovered that pfatp6 allelic variants had a relatively high mutation rate at I898I (33.7%), none of the isolates carried a mutation at pfatp6 S769N. No mutation in the pfk13 gene was observed in any of the studied isolates. A cross-sectional study conducted in Kenya to determine the prevalence of pfk13 substitutions reported that no mutations were associated with ART-resistance in Africa [19]. The present work discovered that pfatp6 S769N, which is known to confer resistance to ART, was wild-type [9]. The absence of mutations in ART-resistance genes indicates that AL has only been in policy for malaria control for 2 years [5].
In conclusion, this study found a high prevalence of pfcrt and pfmdr1 mutations in P. falciparum isolates from 2 endemic areas in southern Somalia, indicating that CQ remains inappropriate for falciparum malaria treatment in Somalia. On the other hand, no mutation in the pfk13 gene was detected. Therefore, AL, which is currently in use in the country, is likely to be effective. However, regular surveillance is essential if the development of widespread ART-resistance is to be prevented.
Acknowledgments
We thank all the patients who participated in the study. Special thanks go to the staff of Balad Hospital and Afgoi General Hospital, especially Mr. Mohamed Omar, Mr. Ahmed Hersi, and Ms. Fartun Mohamed, for their assistance in sample collection. We also thank Chulabhorn International College of Medicine of Thammasat University, Center of Excellence in Pharmacology and Molecular Biology of Malaria and Cholangiocarcinoma of Thammasat University, for the support of this research. Kesara Na-Bangchang is supported by the National Research Council of Thailand under the Research Team Promotion grant (grant number NRCT 820/2563).
Footnotes
We have no conflict of interest related to this work.
Author contributions
Conceptualization: Jalei AA, Chaijaroenkul W
Data curation: Chaijaroenkul W
Formal analysis: Jalei AA, Muhamad P, Chaijaroenkul W
Funding acquisition: Na-Bangchang K
Investigation: Na-Bangchang K
Methodology: Jalei AA, Muhamad P
Resources: Na-Bangchang K, Muhamad P
Supervision: Na-Bangchang K
Validation: Na-Bangchang K, Chaijaroenkul W
Writing – original draft: Jalei AA
Writing – review & editing: Na-Bangchang K, Chaijaroenkul W
References
- 1.Bousema T, Drakeley C. Epidemiology and infectivity of Plasmodium falciparum and Plasmodium vivax gametocytes in relation to malaria control and elimination. Clin Microbiol Rev . 2011;24(2):377–410. doi: 10.1128/CMR.00051-10. https://doi.10.1128/CMR.00051-10 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jalei AA, Chaijaroenkul W, Na-Bangchang K. Plasmodium falciparum drug resistance gene status in the Horn of Africa: a systematic review. Afr J Pharm Pharmacol . 2018;12(25):361–373. doi: 10.5897/AJPP2018.4942. [DOI] [Google Scholar]
- 3.World Health Organization . Guidelines for the Treatment of Malaria. World Health Organization; Geneva, Switzerland: 2015. https://apps.who.int/iris/handle/10665/162441 . [Google Scholar]
- 4.Warsame M, Atta H, Klena JD, Waqar BA, Elmi HH, et al. Efficacy of monotherapies and artesunate-based combination therapies in children with uncomplicated malaria in Somalia. Acta Trop . 2009;109(2):146–151. doi: 10.1016/j.actatropica.2008.10.009. https://doi.10.1016/j.actatropica.2008.10.009 . [DOI] [PubMed] [Google Scholar]
- 5.Warsame M, Hassan AH, Hassan AM, Arale AM, Jibril AM, et al. Efficacy of artesunate+ sulphadoxine/pyrimethamine and artemether+ lumefantrine and dhfr and dhps mutations in Somalia: evidence for updating the malaria treatment policy. Trop Med Int Health . 2017;22(4):415–422. doi: 10.1111/tmi.12847. https://doi.10.1111/tmi.12847 . [DOI] [PubMed] [Google Scholar]
- 6.Ariey F, Witkowski B, Amaratunga C, Beghain J, Langlois AC, et al. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature . 2014;505(7481):50–55. doi: 10.1038/nature12876. https://doi.10.1038/nature12876 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ndwiga L, Kimenyi KM, Wamae K, Osoti V, Akinyi M, et al. A review of the frequencies of Plasmodium falciparum Kelch 13 artemisinin resistance mutations in Africa. Int J Parasitol Drugs Drug Resist . 2021;16:155–161. doi: 10.1016/j.ijpddr.2021.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Djimdé A, Doumbo OK, Cortese JF, Kayentao K, Doumbo S, et al. A molecular marker for chloroquine-resistant falciparum malaria. N Engl J Med . 2001;344(4):257–263. doi: 10.1056/NEJM200101253440403. https://doi.10.1056/NEJM200101253440403 . [DOI] [PubMed] [Google Scholar]
- 9.Jambou R, Legrand E, Niang M, Khim N, Lim P, et al. Resistance of Plasmodium falciparum field isolates to in-vitro artemether and point mutations of the SERCA-type PfATPase6. Lancet . 2005;366(9501):1960–1963. doi: 10.1016/S0140-6736(05)67787-2. [DOI] [PubMed] [Google Scholar]
- 10.Win AA, Imwong M, Kyaw MP, Woodrow CJ, Chotivanich K, et al. K13 mutations and pfmdr1 copy number variation in Plasmodium falciparum malaria in Myanmar. Malar J . 2016;15:1–7. doi: 10.1186/s12936-016-1147-3. https://doi.10.1186/s12936-016-1147-3 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nsanzabana C, Ariey F, Beck HP, Ding XC, Kamau E, et al. Molecular assays for antimalarial drug resistance surveillance: a target product profile. PLoS One . 2018;13(9):e0204347. doi: 10.1371/journal.pone.0204347. https://doi.10.1371/journal.pone.0204347 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shafik SH, Cobbold SA, Barkat K, Richards SN, Lancaster NS, et al. The natural function of the malaria parasite’s chloroquine resistance transporter. Nat commun . 2020;11(1):1–16. doi: 10.1038/s41467-020-17781-6. https://doi.10.1038/s41467-020-17781-6 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Durrand V, Berry A, Sem R, Glaziou P, Beaudou J, et al. Variations in the sequence and expression of the Plasmodium falciparum chloroquine resistance transporter (Pfcrt) and their relationship to chloroquine resistance in vitro. Mol Biochem Parasitol . 2004;136(2):273–285. doi: 10.1016/j.molbiopara.2004.03.016. https://doi.10.1016/j.molbiopara.2004.03.016 . [DOI] [PubMed] [Google Scholar]
- 14.Mwai L, Ochong E, Abdirahman A, Kiara SM, Ward S, et al. Chloroquine resistance before and after its withdrawal in Kenya. Malar J . 2009;8:1–10. doi: 10.1186/1475-2875-8-106. https://doi.10.1186/1475-2875-8-106 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ikegbunam MN, Nkonganyi CN, Thomas BN, Esimone CO, Velavan TP, et al. Analysis of Plasmodium falciparum Pfcrt and Pfmdr1 genes in parasite isolates from asymptomatic individuals in Southeast Nigeria 11 years after withdrawal of chloroquine. Malar J . 2019;18(1):1–7. doi: 10.1186/s12936-019-2977-6. https://doi.10.1186/s12936-019-2977-6 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Veiga MI, Dhingra SK, Henrich PP, Straimer J, Gnädig N, et al. Globally prevalent PfMDR1 mutations modulate Plasmodium falciparum susceptibility to artemisinin-based combination therapies. Nat Commun . 2016;7:1–12. doi: 10.1038/ncomms11553. https://doi.10.1038/ncomms11553 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kavishe RA, Paulo P, Kaaya RD, Kalinga A, van Zwetselaar M, et al. Surveillance of artemether-lumefantrine associated Plasmodium falciparum multidrug resistance protein-1 gene polymorphisms in Tanzania. Malar J . 2014;13:1–6. doi: 10.1186/1475-2875-13-264. https://doi.10.1186/1475-2875-13-264 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ngalah BS, Ingasia LA, Cheruiyot AC, Chebon LJ, Juma DW, et al. Analysis of major genome loci underlying artemisinin resistance and pfmdr1 copy number in pre-and post-ACTs in western Kenya. Sci Rep . 2015;5:1–6. doi: 10.1038/srep08308. https://doi.10.1038/srep08308 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muwanguzi J, Henriques G, Sawa P, Bousema T, Sutherland CJ, et al. Lack of K13 mutations in Plasmodium falciparum persisting after artemisinin combination therapy treatment of Kenyan children. Malar J . 2016;15:1–6. doi: 10.1186/s12936-016-1095-y. https://doi.10.1186/s12936-016-1095-y . [DOI] [PMC free article] [PubMed] [Google Scholar]