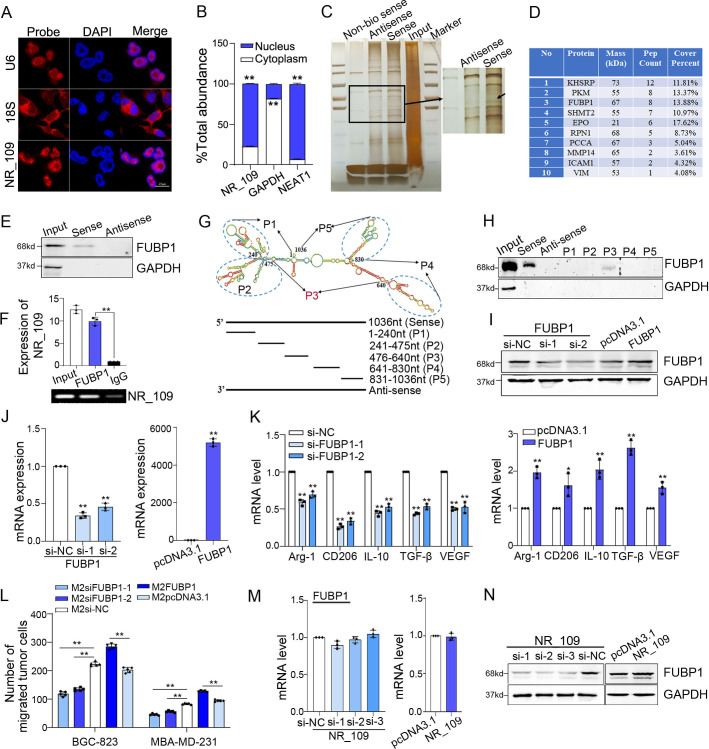
Figure 3.
NR_109 interacted with FUBP1 protein. (A) FISH and (B) nuclear and cytosolic RNA analyses revealed that NR_109 was mainly located in the nucleus of M2-like macrophages. (C) The interacted proteins of NR_109 were resolved through NR_109 pulldown assay, SDS-PAGE electrophoresis and visualized by silver staining. (D) The top 10 proteins analyzed by MS were shown. (E) Only biotinylated sense probe pulled down FUBP1 in M2-like macrophages. (F) The interaction of NR_109 and FUBP1 was confirmed by RIP assays. (G) A series of truncated probes of NR_109 was designed according to the secondary structure of NR_109.(H) The FUBP1 protein was pulled down by the P3 probe (476-640nt) of NR_109. (I-J) M2-FUBP1low and M2-FUBP1high macrophages were constructed by transfecting FUBP1 siRNA or FUBP1 plasmid, respectively. (K) Expression of M2 markers and (L) the migration of tumor cells were measured by qPCR and transwell assays. (M-N) NR_109 affected FUBP1 at the protein level, but not the mRNA level. The statistical data are from three independent experiments and the bar indicates the SD values (*p < 0.05, **p < 0.01). FUBP1, far upstream element-binding protein 1.