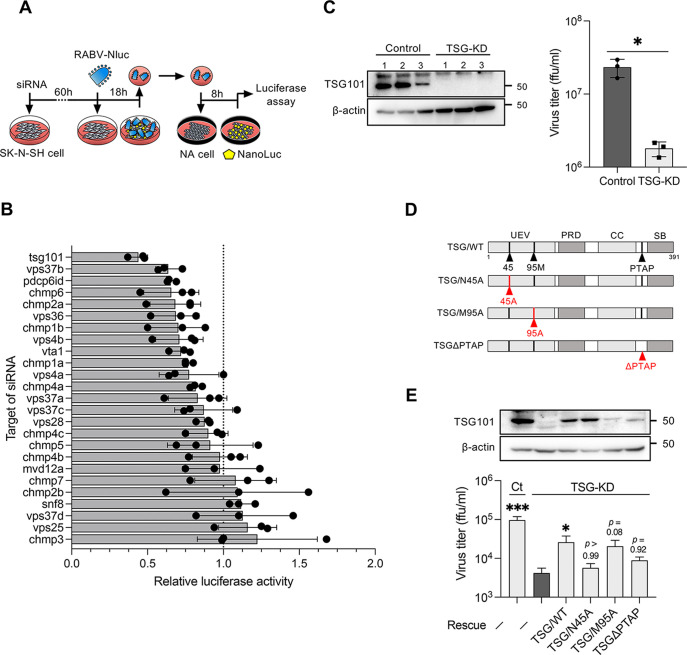
FIG 1.
RNAi screening identifies TSG101 as an ESCRT factor supporting RABV infection. (A) Schematic image of the RNAi screening method. siRNA-transfected SK-N-SH cells were infected with RABV-Nluc at an MOI of 10, and culture supernatants collected at 18 hpi were passaged into NA cells. Luciferase activity in the NA cells was measured at 8 hpi. (B) Luciferase activity derived from NanoLuc-encoded reporter RABV in siRNA-treated SK-N-SH cells relative to the luciferase activity in control siRNA-treated cells. Dots indicate the mean of three different siRNAs for each target. Bars indicate the means ± standard deviations of the three siRNAs. (C) Virus titers in the supernatants of TSG-KD SK-N-SH cells at 48 hpi. siRNA-treated cells were infected with RABV at an MOI of 1. The titers were measured using a focus-forming assay. Bars indicate the means ± standard deviations of three replicates from a representative experiment. (D) Schematic images of the TSG101 mutants used in this study. Mutation sites are marked in red. UEV, ubiquitin-conjugating enzyme E2 variant; PRD, proline-rich domain; CC, coiled-coil domain; PTAP, conserved PTAP tetrapeptide motif; SB, steadiness box. (E) Virus titers in TSG-KD and rescue cells. TSG-KD cells were transfected with siRNA-resistant TSG101-encoding plasmids and infected with RABV at an MOI of 1. The virus titers in supernatants at 24 hpi were measured. Bars indicate the means ± standard deviations of three replicates from a representative experiment. For statistical analyses, Welch’s t test used in panel C (*, P < 0.05), and one-way ANOVA and Dunn’s multiple-comparison tests were used in panel E (*, P < 0.05; ***, P < 0.001).