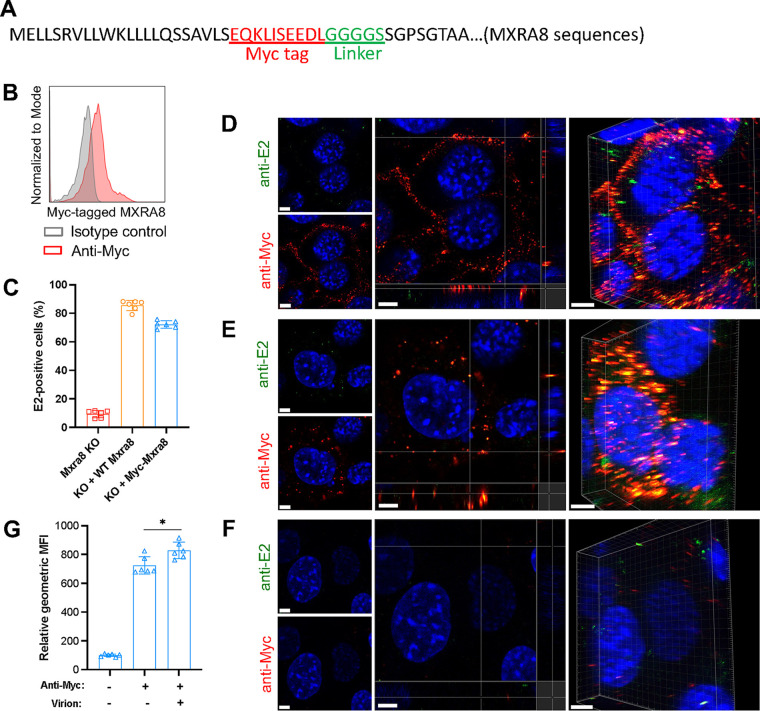
FIG 5.
Myc-tagged MXRA8 colocalizes with CHIKV virions. ΔMxra8 MEFs were complemented with Myc-tagged MXRA8. (A) Insertion of a Myc tag plus a linker in MXRA8. The amino acid sequence is indicated. (B) The expression of Myc-tagged MXRA8 in ΔMxra8 MEFs. The complemented cells were stained with anti-Myc tag or isotype control antibody and were subjected to flow cytometry analysis. (C) Functional assessment of Myc-tagged MXRA8 in ΔMxra8 MEFs. The complemented cells were infected with CHIKV for 8 h, followed by E2 antigen detection using flow cytometry. Data shown are an average of two independent experiments performed in triplicate (mean ± SD). (D) Myc-MXRA8 colocalizes with CHIKV virions at the cell surface. Cells were incubated with virus particles at 4°C for 30 min and incubated with anti-E2 and anti-Myc primary antibodies at 4°C for 30 min. After washing, cells were fixed and stained with fluorophore-labeled secondary antibodies. Orthogonal and 3D views of colocalization are presented. (E) Myc-MXRA8 colocalizes with CHIKV particles in the cytoplasm. Cells were incubated with viral particles and antibodies at 37°C for 15 min and subsequently placed on ice. Surface virions and antibodies were removed by treatment with protease K. Cells were fixed, permeabilized, and stained with fluorophore-labeled secondary antibodies. Orthogonal and 3D views of colocalization are presented. (F) As a control, cells before fixation were treated with proteinase K to remove bound virus particles and antibodies, followed by fixation and surface staining with secondary antibodies to monitor stripping efficiency. In D to F, one representative image from at least three independent experiments is shown; scale bar, 5 μm. (G) Myc-tagged MXRA8 proteins were labeled with anti-Myc antibody to assess the internalization efficiency in the presence or absence of virions. Cells in suspension were processed similar to as shown in E and analyzed by flow cytometry. The geometric mean fluorescence intensity (GMFI) of anti-Myc antibody-positive cells was measured. Data shown are an average of two independent experiments performed in triplicate. Data were analyzed by unpaired t test and are presented as mean ± SD; *, P < 0.05.