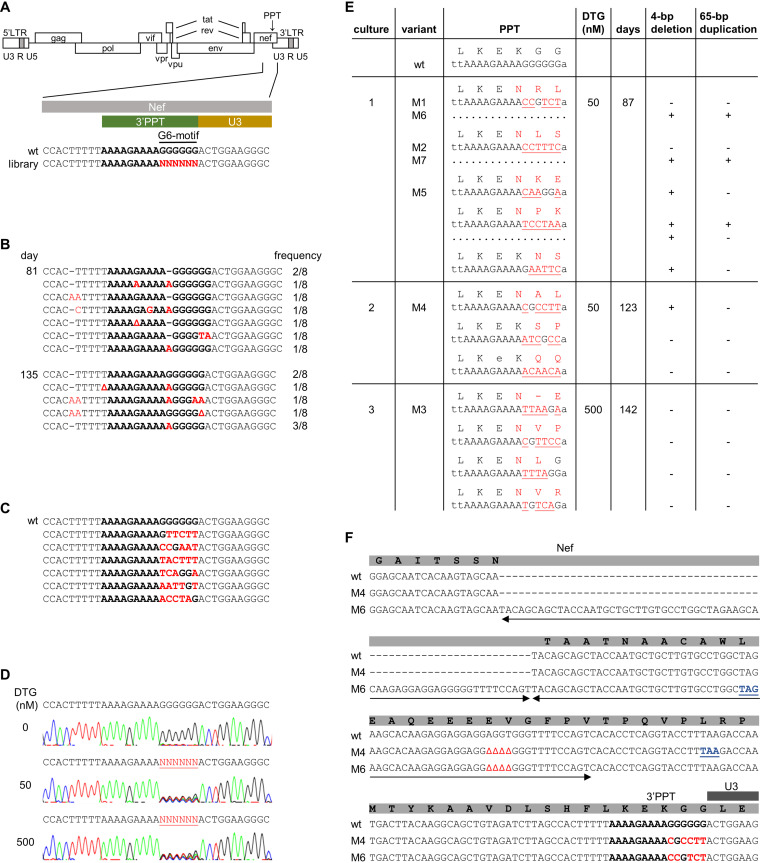
FIG 2.
Selection of DTG-resistance mutations in the HIV 3’PPT. (A) Schematic of the HIV genome and nucleotide sequence of the 3′PPT region. The 3′PPT nucleotide sequence is indicated in bold. The G6-motif that is randomized in the SELEX library is indicated in red. (B) 3′PPT-mutations identified upon long-term culturing of HIV with DTG on SupT1 T cells. After 81 and 135 days of virus passage, intracellular DNA was isolated and the HIV 3′PPT region was amplified by PCR, followed by sequencing of 8 individual PCR clones. The frequency by which each 3′PPT sequence was found is indicated (Δ, nucleotide deletion; dashes are used to align sequences). The 3′PPT nucleotide sequence is indicated in bold. Nucleotide substitutions are in red. (C) The 3′PPT sequence in individual plasmid clones from the full-length HIV-1 library in which the G6-motif was randomized. The wt sequence is shown on top for comparison. The 3′PPT nucleotide sequence is indicated in bold. Nucleotide substitutions are in red. (D) C8166 T cells were infected with the HIV-1 library and cultured with 0, 50, or 500 nM DTG. Upon syncytium formation (at day 7 without DTG and day 14 with DTG), the 3′PPT region of the virus population was PCR amplified and analyzed by sequencing. The randomized G6-motif is indicated in red and underlined. (E) Sequence analysis of the 3′PPT/nef region upon long-term culturing of the HIV-1 library with 50 (cultures 1 and 2) or 500 nM DTG (culture 3). Intracellular viral DNA was isolated from the infected C8166 cells at the indicated day, followed by PCR amplification of the 3′PPT/nef region and ligation of the PCR product into a TOPO TA cloning vector. Several TA clones were sequenced. The observed 3'PPT nucleotide sequences with their amino acid translation of the overlapping nef open reading frame are shown. Nucleotide substitutions are in red and underlined. Amino acid substitutions are in red. When indicated, a 4-bp deletion and 65-bp duplication (sequence shown in F) was detected in the upstream nef region. The variants indicated with M1-M7 were cloned into the full-length HIV-1 LAI construct for further analysis. (F) Sequence alignment of the 3′PPT/nef region in wt HIV and the M4 and M6 variants. The 3′PPT nucleotide sequence is indicated in bold. Nucleotide substitutions are indicated in red, deletions are indicated by Δ, and the duplicated sequence is indicated with two-sided arrows. The deletion and duplication result in a premature stop codon in the nef open reading frame (in blue and underlined).