Fig. 1.
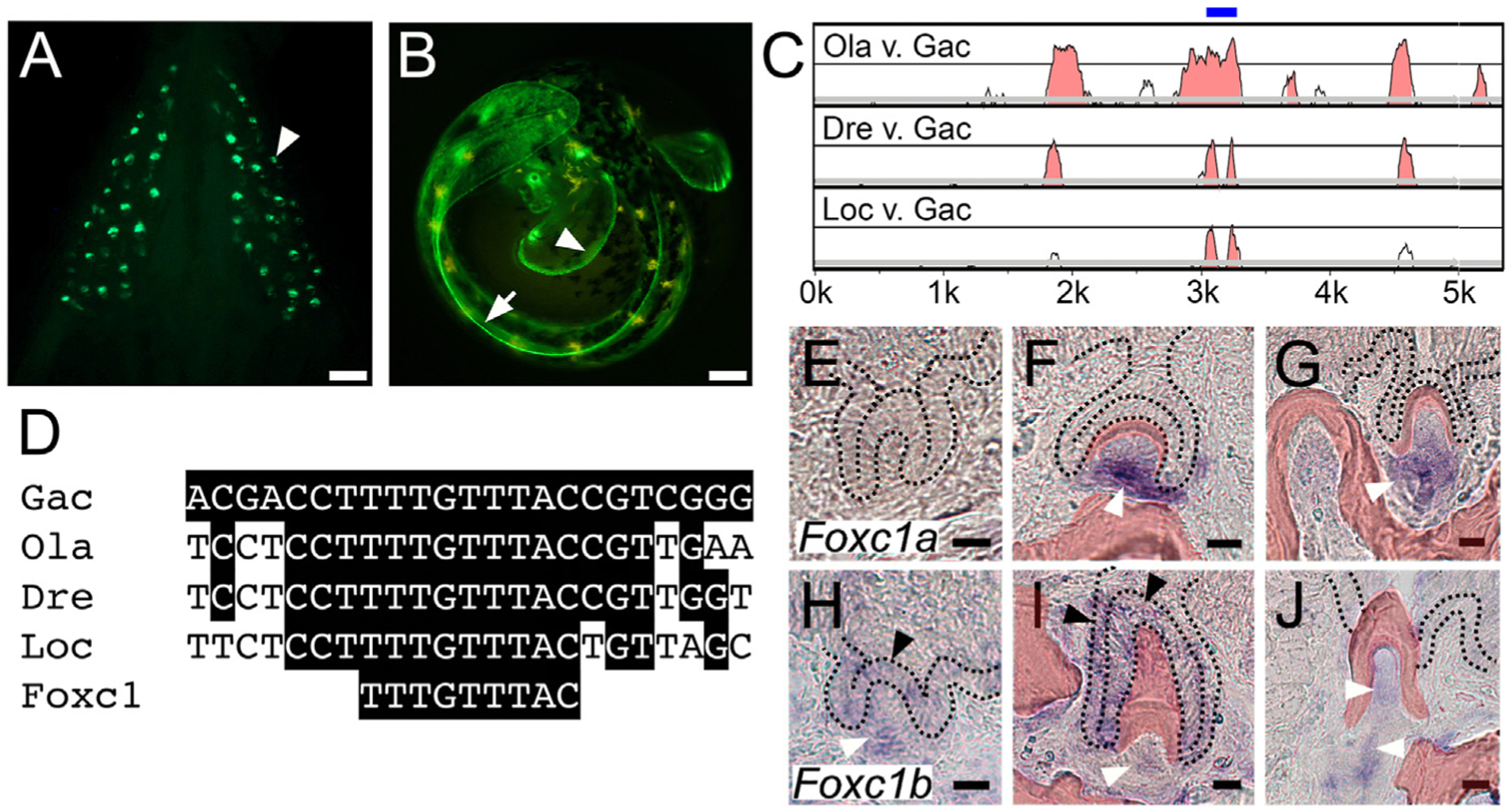
A pleiotropic tooth and fin enhancer includes an evolutionarily conserved predicted Foxc1 binding site and Foxc1 genes are expressed in teeth. GFP reporter gene driven by a 1.3 kb Bmp6 intronic enhancer (Cleves et al., 2018; Stepaniak et al., 2021) reveals expression in (A) ventral pharyngeal teeth (arrowhead) of juvenile stickleback fish and (B) distal edge of pectoral (arrowhead) and median (arrow) fin fold of unhatched 7 dpf developing stickleback embryo. (C) mVISTA LAGAN sequence alignment showing central region of stickleback Bmp6 intron four is conserved to gar. Intron four of stickleback Bmp6 (x-axis) is aligned to medaka (top), zebrafish (middle) and gar (bottom). Graph shows a sliding window of 100 base pairs covering the first ~5.5 kb of this ~7 kb intron, with y-axis starting at 50% identity and windows shaded at a threshold of 70% nucleotide identity. The blue bar above the peaks indicates the central ~200 bp island of conservation (see Fig. S1). Gac = Gasterosteus aculeatus (stickleback), Ola = Oryzias latipes (medaka), Dre = Danio rerio (zebrafish), Loc = Lepisosteus oculatus (gar). (D) Predicted Foxc1 binding site TTTGTTTAC (Berry et al., 2008) within the central island of conservation is conserved from sticklebacks to gar. In situ hybridization of Foxc1a (E–G) and Foxc1b (H–J) expression on sections of adult stickleback pharyngeal teeth at three different stages: mid bell (E,H), late bell (F,I), and erupted (G,J). Foxc1a expression was detected in dental mesenchyme at late bell and erupted stages (white arrowheads), but not at mid bell. Foxc1b expression was detected in tooth germ epithelium at mid bell (black arrowhead in H) and in both the inner (left black arrowhead) and outer dental epithelium (top black arrowhead) at late bell (I). Foxc1b was also detected in tooth mesenchyme at all three stages (white arrowheads, H-J). Black dotted lines demarcate the dental epithelium, and bone and dentine is pseudocolored red (see Fig. S2 for unlabeled panels and DAPI staining). Scale bars = 10 μm.
