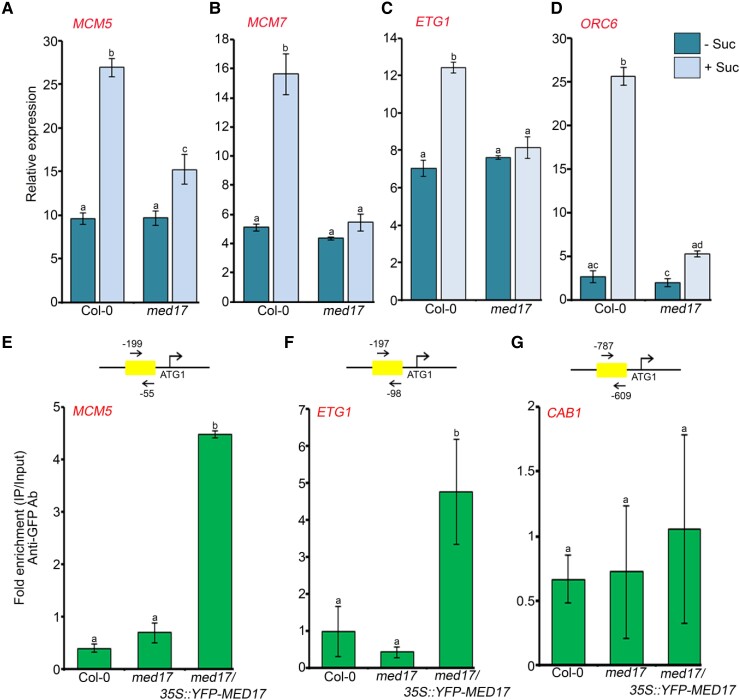
Figure 4.
MED17 regulates the expression of cell cycle genes during root meristem activation. A–D, RT-qPCR showing expression of cell cycle genes (MCM5/7, ETG1, and ORC6) after sucrose treatment. Four-day-old Col-0 and med17 seedlings were treated with 90 mM sucrose for 3 h. Gene expression values were calculated as ΔCt. RT-qPCR analysis was performed on three independent biological replicates (n = 3). Bar plots represent mean values, and error bars denote Se. Statistical difference has been depicted by P-value as assessed by one-way ANOVA and Tukey's HSD post hoc test. E–G, ChIP-qPCR showing enrichment of YFP-MED17 at the promoters of MCM5, ETG1, and CAB1 in 7-day-old Col-0, med17, and med17/35S:YFP-MED17 seedlings. The promoter regions harboring the E2F-binding elements were amplified. Amplicon positions relative to ATG (translation start site) are shown in the upper panel. Ct values with and without antibody samples were normalized to input control. Untransformed Col-0 and med17 seedlings were taken as negative control. CAB1, which does not possess any E2F-binding motifs in its promoter, was used as a negative control. YFP-MED17 binding on the promoter regions was calculated as fold enrichment. ChIP-qPCR analysis was performed on three technical replicates (n = 3) from a single representative experiment. Experiments were independently repeated twice (biological replicates; n = 2). Bar plots represent mean values, and error bars denote Sd. Statistical difference has been depicted by P-value as assessed by one-way ANOVA and Tukey's HSD post hoc test. For all the graphs, P-value of 0.05 or lower (P ≤ 0.05) was considered statistically significant, whereas P-value greater than 0.05 (P > 0.05) was considered non-significant (ns).