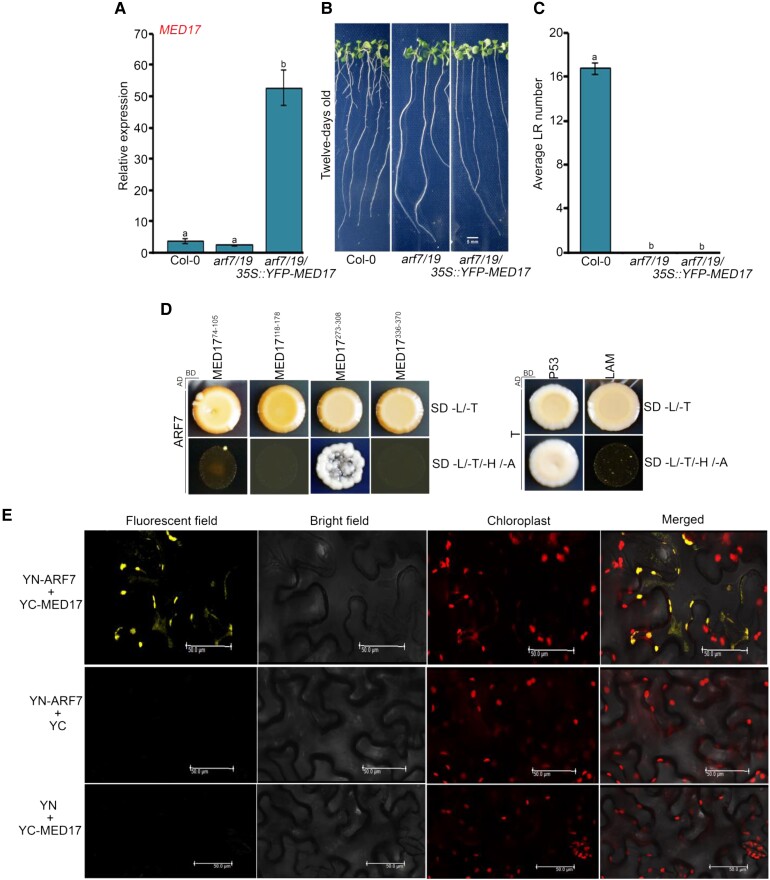
Figure 6.
MED17 regulates LR development in an ARF-dependent manner. A, RT-qPCR showing the expression level of MED17 in Col-0, arf7/19, and arf7/19/35S:YFP-MED17 seedlings. The seedlings were grown on ½ MS for 7 days and then harvested for RNA isolation. Gene expression values were calculated as ΔCt. RT-qPCR analysis was performed on three independent biological replicates (n = 3). Bar plots represent mean values, and error bar denotes Se. Statistical difference has been depicted by P-values as assessed by one-way ANOVA and Tukey's HSD post hoc test. B, Root phenotype of Col-0, arf7/19, and arf7/19/35S::YFP-MED17 seedlings. Length of the scale bar in the image is 5 mm. C, Graphical representation of the average number of lateral roots (LRs) per seedling in Col-0, arf7/19, and arf7/19/35S::YFP-MED17. Data shown are the average of three independent biological replicates containing at least 20 seedlings. Bar plots represent mean values, and error bar denotes Se. Statistical difference has been depicted by P-values as assessed by one-way ANOVA and Tukey's HSD post hoc test. D, Y2H assay to study the interaction of MED17 with ARF7. Interactions were scored by growth of yeast colonies on quadruple drop out (QDO) (-H/-L/-T/-A) selection media. P53-T and LAM-T were used as positive and negative controls, respectively. E, BiFC assay showing interaction of MED17 with ARF7. MED17 and ARF7 were cloned into CD3-1651 and CD3-1648 vectors, respectively. Interaction was confirmed on the basis of YFP signals. No YFP signals were observed between YFP N ARF7–C YFP and YFP N–C YFP MED17. Hence, they served as negative controls. Length of the scale bar in the image is 50 µm. For all the graphs, P-value of 0.05 or lower (P ≤ 0.05) was considered statistically significant, whereas P-value greater than 0.05 (P > 0.05) was considered non-significant (ns).