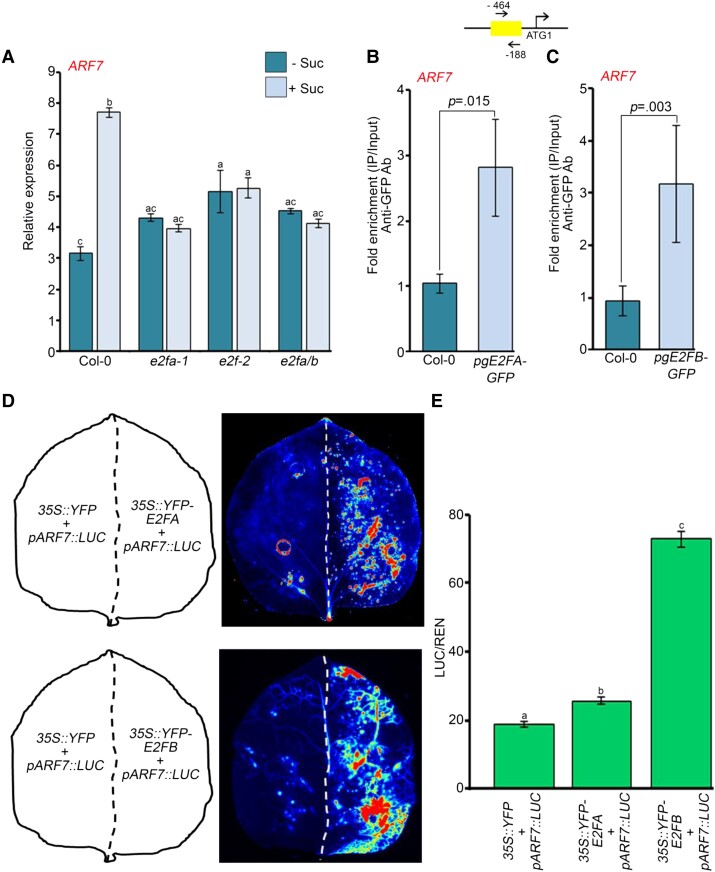
Figure 7.
E2F TFs are required for sucrose-induced expression of ARF7. A, RT-qPCR showing expression of ARF7 in Col-0, e2fa-1, e2fb-2, and e2fa/b seedlings after sucrose treatment. Seven-day old Col-0, e2fa-1, e2fb-2, and e2fa/b seedlings were treated with 90 mM sucrose for 3 h. Gene expression values were calculated as ΔCt. RT-qPCR analysis was performed on three independent biological replicates (n = 3). Bar plots represent mean values, and error bars denote Se. Statistical difference has been depicted by P-value as assessed by one-way ANOVA and Tukey's HSD post hoc test. B, ChIP-qPCR showing enrichment of E2FA–GFP at the promoter of ARF7 in 7-day-old Col-0 and pgE2FA-GFP seedlings. C, ChIP-qPCR showing enrichment of E2FB–GFP at the promoter of ARF7 in 7-day-old Col-0 and pgE2FB-GFP seedlings. In (B and C), the promoter region of ARF7 harboring E2F-binding element was amplified. Ct values with and without antibody samples were normalized to input control. Untransformed Col-0 seedlings were taken as negative control. E2FA–GFP and E2FB–GFP binding on promoters were calculated as fold enrichment. ChIP-qPCR analysis was performed on three technical replicates (n = 3) from a single representative experiment. Experiments were independently repeated twice (biological replicates; n = 2). Bar plots represent mean values, and error bars denote Sd. Statistical difference has been depicted by P-value as assessed by paired t test. D, Luciferase activity assay for determining the activation of ARF7 promoter by E2FA and E2FB TFs. Luciferase was cloned under the control of ARF7 promoter. The left halves of the leaves were infiltrated with 35S::YFP and pARF7::LUC plasmids whereas the right halves of the leaves were infiltrated with either 35S::YFP-E2FA or 35S::YFP-E2FB plasmid along with pARF7::LUC plasmid. Activation of ARF7 promoter by E2FA or E2FB was assessed by measuring the luciferase signals. E, Graph showing the quantification of luminescence from pARF7::LUC in the presence or absence of E2FA/B. Relative luminescence was calculated by dividing luminescence of firefly luciferase by luciferase of Renilla. Experiments were independently repeated twice (biological replicates; n = 2, each containing three technical replicates). Bar plots represent mean values, and error bars denote Se. Statistical difference has been depicted by P-value as assessed by one-way ANOVA and Tukey's HSD post hoc test. For all the graphs, P-value of 0.05 or lower (P ≤ 0.05) was considered statistically significant, whereas P-value greater than 0.05 (P > 0.05) was considered non-significant (ns).