Figure 1: Data are collected, processed by the T2DKP platform, and provided through the T2DKP web-interface via a multi-step process.
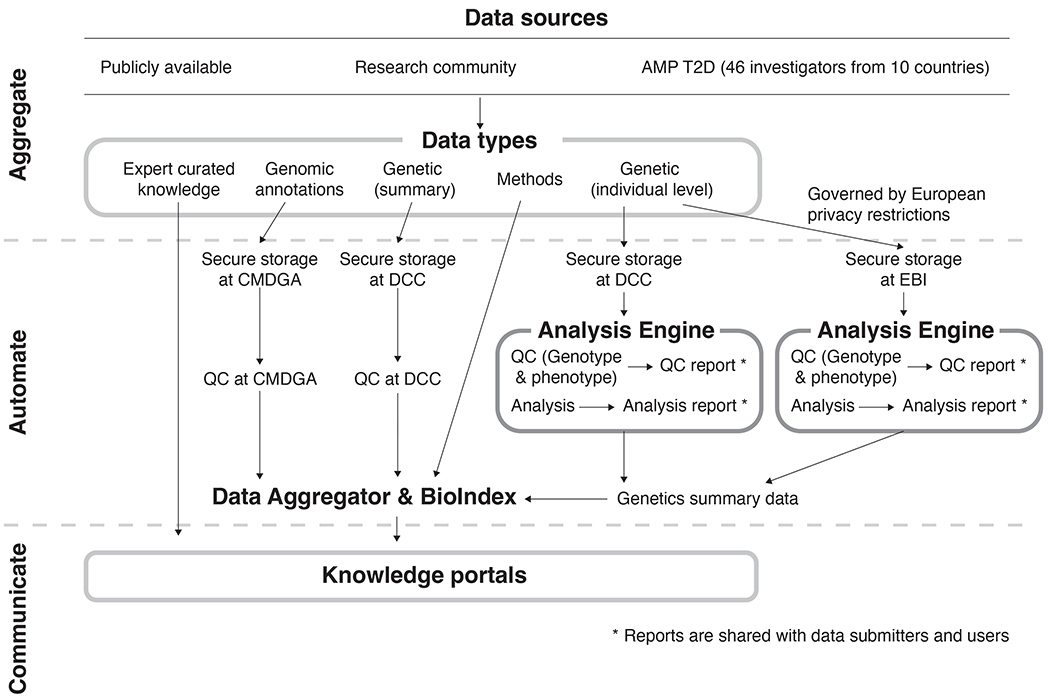
Data sources for the T2DKP are of varied origin and of multiple Data types. Summary-level genetic datasets are transferred to the Data Coordinating Center (DCC) at the Broad Institute, while genomic annotations are transferred to the Common Metabolic Diseases Genome Atlas (CMDGA). Individual-level genetic datasets are transferred to the DCC or European Bioinformatics Institute (EBI) depending on permissions, and the Analysis Engine processes them through a common analytical workflow to produce summary-level associations. The Data Aggregator then analyzes summary-level genetic datasets and genomic annotations with a series of bioinformatic methods, the results of which are stored in the BioIndex. The Knowledge portals access the data within the BioIndex and present them via a web-interface.
