Abstract
Aims
One of the most important complications of heart transplantation is organ rejection, which is diagnosed on endomyocardial biopsies by pathologists. Computer-based systems could assist in the diagnostic process and potentially improve reproducibility. Here, we evaluated the feasibility of using deep learning in predicting the degree of cellular rejection from pathology slides as defined by the International Society for Heart and Lung Transplantation (ISHLT) grading system.
Methods and results
We collected 1079 histopathology slides from 325 patients from three transplant centres in Germany. We trained an attention-based deep neural network to predict rejection in the primary cohort and evaluated its performance using cross-validation and by deploying it to three cohorts. For binary prediction (rejection yes/no), the mean area under the receiver operating curve (AUROC) was 0.849 in the cross-validated experiment and 0.734, 0.729, and 0.716 in external validation cohorts. For a prediction of the ISHLT grade (0R, 1R, 2/3R), AUROCs were 0.835, 0.633, and 0.905 in the cross-validated experiment and 0.764, 0.597, and 0.913; 0.631, 0.633, and 0.682; and 0.722, 0.601, and 0.805 in the validation cohorts, respectively. The predictions of the artificial intelligence model were interpretable by human experts and highlighted plausible morphological patterns.
Conclusion
We conclude that artificial intelligence can detect patterns of cellular transplant rejection in routine pathology, even when trained on small cohorts.
Keywords: Computational pathology, Artificial intelligence, Self-supervised deep learning, Multiple instance learning, Cardiac allograft rejection
Graphical Abstract
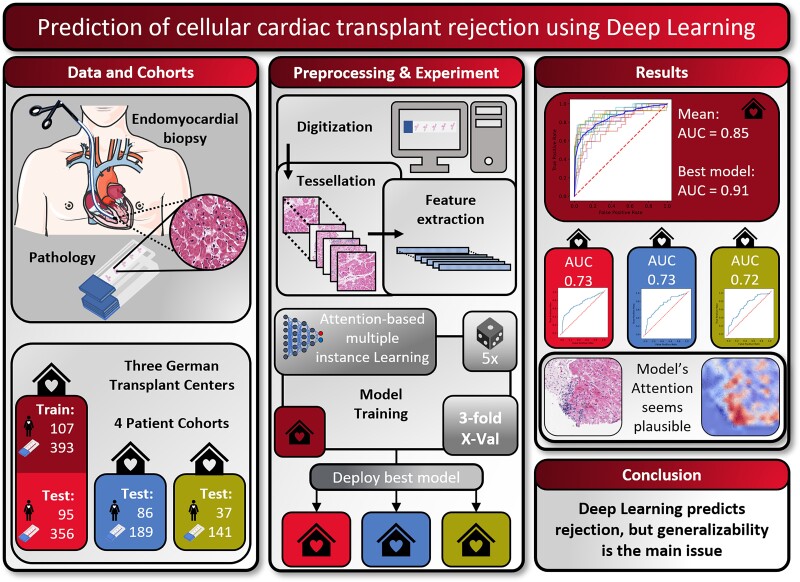
Graphical Abstract.
Introduction
In patients with end-stage heart failure, organ transplantation constitutes the desired curative treatment concept.1 This has been made possible in recent decades, in particular, by the advent of new immunosuppressive drugs, which can ensure long-lasting organ preservation. However, organ rejection by the host immune system remains one of the major complications in these patients.2 Despite the increasing importance of noninvasive methods in the detection of graft rejection, endomyocardial biopsy remains the gold standard for detecting rejection, especially in the first year after transplantation.3,4 The pathological assessment of such specimens is reserved for highly specialized pathologists and has massive clinical consequences. In 1990, the International Society for Heart and Lung Transplantation (ISHLT) published a guideline for histopathologic diagnosis of acute cellular rejection to standardize this assessment, which has been revised in 2004.5 Nevertheless, the purely subjective assessment of pathological sections has certain disadvantages, such as the dependency on appropriately trained experts, as well as remaining inter- and intra-observer variability.6 In addition, endomyocardial biopsies are obtained either as routine surveillance protocol diagnostics or as a diagnostic investigation in patients with allograft dysfunction and clinically suspected rejection.
Computer-based image analysis programmes can potentially support pathology experts in performing diagnostics. In several histopathological applications, it could be shown that such computer-based image analysis programmes can show a high level of concordance with human observers, and in some cases, the combination with the human experts can improve the consistency of the findings.7 In particular, the technology of artificial neural networks has brought very good results in many clinically relevant prediction tasks in recent years.8,9 A recent extension of this technology is the so-called attention-based multiple instance learning (MIL),10 in which the artificial neural network can learn which areas of the whole slide image are more relevant than other areas.11,12
In contrast to solid tumours, in which many studies have examined computer-based prediction of clinically relevant biomarkers in the last 3 years,9 there are only comparatively few studies in the context of transplantation medicine. Precedent cases exist in the prediction of organ rejection after kidney transplantation,13 as well as applications of simple, handcrafted feature-based image analysis methods to cardiac biopsies after transplantation.14,15 Handcrafted features may provide a benefit in interpretability and are well established, but in almost all applications of computational pathology, deep learning methods are emerging as more powerful and more versatile methods.11,16 A recent study by Lipkova et al.17 used the deep learning pipeline ‘CRANE’ to predict cardiac allograft transplantation, yielding a very high and clinical-grade performance.18
However, several open questions remain regarding the data requirements to train such systems, as Lipkova et al. trained their system on thousands of patient samples, but this large number of samples is rarely available. Additional questions remain open regarding the generalizability of such systems and the biological interpretability which can be drawn from their predictions. Finally, new technical approaches such as self-supervised learning (SSL) to pre-train pathology deep learning models could yield an improved performance,19 but this has not yet been evaluated in the prediction of cardiac allograft rejection.
In the present study, we collected four cohorts from three hospitals of cardiac transplant patients undergoing cardiac biopsy routinely and based on clinically relevant changes. We trained our own deep learning pipeline using an SSL-based feature extractor combined with attention-based MIL and compared the performance to the CRANE method for the prediction of cellular transplant rejection in a multicentric data set.
Methods
Patient cohorts and experimental design
In this study, we included four case series (‘patient cohorts’) from three different medical centres in Germany. The first cohort was obtained from the pathological archive of the University Hospital Regensburg and contained 393 pathological sections from 107 patients from the period 2016 to 2018. The second cohort also originated from the pathological archive of the University Hospital Regensburg and contained 356 pathological sections from 95 patients from the period 2019 to 2021. The third cohort was obtained from the pathological archive of the University Medical Center Hamburg-Eppendorf. This cohort contained 189 pathological sections from 86 patients from the period 2019 to 2021. The fourth cohort was obtained from the pathological archive of the University Hospital Aachen containing 141 pathological sections from 37 patients from the period 1999 to 2014. Cohorts were consecutive retrospective case series. We did not perform any formal sample size calculation, but rather pragmatically aimed to maximize the sample size of training and testing cohorts. The ground truth was obtained by two expert pathologists during routine work-up at each participating centre, grading the degree of rejection in consensus, following the 2004 revision of the ISHLT grading system.5 All patient samples without information on ISHLT grading were not eligible for inclusion. A detailed presentation of the clinical characteristics of all patients in the corresponding cohorts can be found in Table 1. We used two categorizations of the ISHLT 2004 grading system as our prediction target. The first is a binarized target (ISHLT 2004 rejection ‘yes/no’), summarizing slides with ISHLT 0R on the one hand (class ‘no’) and all signs of rejection on the other hand (ISHLT 1R, 2R, 3R; class ‘yes’). For the second target (‘ISHLT 2004 rejection grade’), we aimed for a more granular classification splitting the second class giving three classes comprising ISHLT 0R, ISHLT 1R, and ISHLT 2R and 3R. We combined the higher order rejection due to shortage of ISHLT 3R cases in the training set (Table 1). Our study adheres to the STARD guidelines (see Supplementary material online, Table S1).20
Table 1.
Clinical characteristics of all cohorts
| Cohort 1 | Cohort 2 | Cohort 3 | Cohort 4 | |
|---|---|---|---|---|
| Contributing centre | Regensburg | Regensburg | Hamburg | Aachen |
| Use in this study | Train | Test | Test | Test |
| N patients | 107 | 95 | 86 | 37 |
| N slides | 393 | 356 | 189 | 141 |
| Recruitment years | 2016–18 | 2019–21 | 2019–21 | 1999–2014 |
| Age at biopsy in years (median, IQR) | 57 (12) | 59 (13.5) | 52 (17) | 55 (12) |
| Gender (F:M per slides) | 55:338 | 55:301 | 70:121 | 47:94 |
| ISHLT rejection | ||||
| No | 312 | 271 | 130 | 84 |
| Yes | 81 | 85 | 59 | 57 |
| ISHLT 0R | 312 | 271 | 130 | 57 |
| ISHLT 1R | 51 | 77 | 51 | 24 |
| ISHLT 2/3R | 30 | 8 | 8 | 60 |
Sample processing and image pre-processing
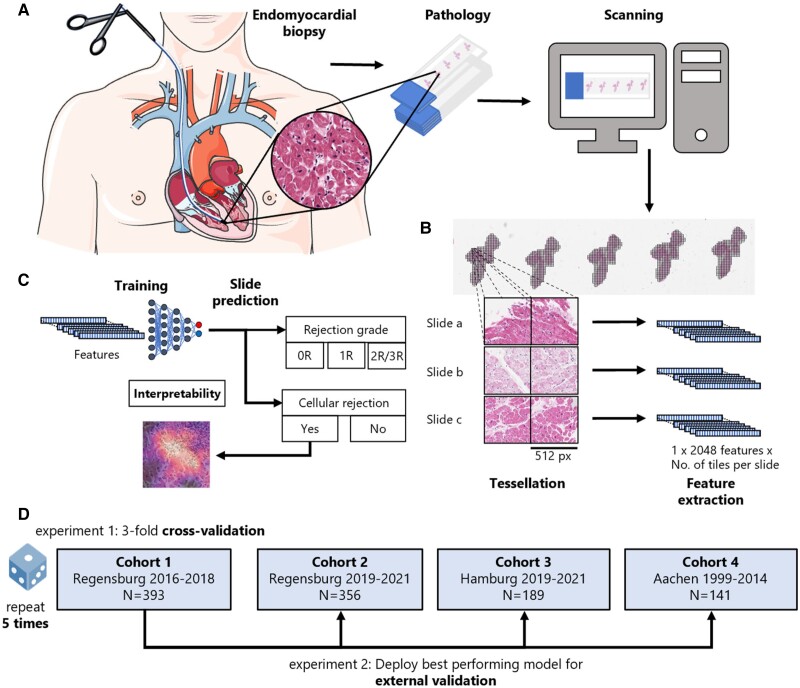
Routine tissue sections were obtained from the pathology archives at the above-mentioned institutions. All slides were stained with haematoxylin and eosin (H&E) according to the standard clinical protocols at each centre. Pen marks were removed from the slides of the training cohort. All images were digitized at ×40 magnification with an Aperio AT2 Slide scanner (Aperio, Leica Camera AG, Wetzlar, Germany) centrally at the University Hospital Düsseldorf (Figure 1A). All images were available in ScanScope Virtual Slide (SVS) format and were tessellated in tissue patches of 512 × 512 pixel size using https://github.com/KatherLab/preprocessing-ng according to the ‘The Aachen Protocol for Deep Learning Histopathology: A hands-on guide for data preprocessing’ (Figure 1B).21
Figure 1.
Outline of the study procedures. (A) Routine endomyocardial biopsies of the right interventricular septum were taken from heart-transplanted patients. These biopsies were then prepared into H&E-stained histopathology slides, before being digitized and turned into whole slide images (WSIs) by use of a slide scanner (icon from smart.servier.com). (B) To make these WSIs processable for our attention-based deep learning models, in a first step, they need to be cut into smaller tiles while the background and artefacts are removed (tessellation). In the next step, feature maps are extracted from all tiles from all slides using a publicly available neural network, which has been pre-trained by self-supervised learning with thousands of histopathology images. (C) The resulting bags of feature maps per slide, together with expert pathologists’ opinion on the occurrence of rejection on a slide level as target label, are then used as training input for an attention-based deep learning model. (D) In a first experiment, three-fold cross-validation is performed within Cohort 1 and repeated five times. In a second experiment, the best performing model from Experiment 1 is externally validated on Cohorts 2, 3, and 4.
Deep learning workflow
For all deep learning experiments, we used our in-house pipeline ‘Marugoto’, which is publicly available at https://github.com/KatherLab/marugoto and has been previously used for analysis of images obtained from cancer tissue.22 In this approach, each image tile was translated into a 2048-dimensional feature vector by a pre-trained histology-specific encoder RetCCL (https://github.com/Xiyue-Wang/RetCCL).23 This encoder has been pre-trained on a large data set of histopathology images with clustering-guided contrastive learning. We used attention-based multiple instance learning,10 in which all feature vectors obtained from all tiles from one whole slide image constitute a ‘bag’ which is processed by the neural network (Figure 1B). The multiple instance learning network is structured as follows: The feature vectors of each of the bag’s tiles are first projected into a length 256 feature space using a fully connected layer. Based on these, an attention module consisting of two fully connected layers calculates an attention score for each of the tiles. All of a bag’s attention scores are then normalized using softmax. We then calculate a bag-level feature vector by taking the sum of the tiles’ feature projections weighted by their respective attention scores. The final classification is then done with an additional fully connected layer (Figure 1C, Supplementary material online, Figure S1). During training, we limited our bag size to a maximum of 512 tiles from each slide, randomly resampled in each epoch (median number of tiles = 403, interquartile range = 468). For slides containing less than 512 tiles, we padded with zeros. During training, we validated the model’s performance at each epoch on the validation data set using all available tiles from each slide. For training, we used an optimal learning rate finder provided by the Python library ‘fastai’ (learner.lr_find). We used the Adam optimizer during training.24 We stopped the training of our model if no reduction in the validation loss was present for the 16 following epochs while training for a maximum of 32 epochs. For deployment, we used all of the slides’ tiles. We compared our approach to CRANE as presented by Lipkova et al.17 To do so, we followed the workflow of the CRANE study, pre-processing the slides with the CLAM repository, which uses a ResNet50 pre-trained on ImageNet as a feature encoder and performed 10-fold Monte-Carlo cross-validation on our training cohort, deploying the best performing model on our test cohorts.25
Experimental design and hardware
We pre-specified the following experimental design. First, we trained and evaluated our SSL-attention algorithm in the first cohort via three-fold cross-validation and repeated this experiment five times. Specifically, we used the sklearn.model_selection.StratifiedKFold() function of sklearn with n_splits = 3 forming three equally sized splits with equal distribution of classes. Accordingly, two parts of the cohort were used for training and validation while the third part was used as a test cohort within this cross-validation experiment. To generate the validation set from the two cohort parts used for training, we applied the sklearn.model_selection.train_test_split function, which by default splits these parts randomly into a training set of 75% and a validation set of 25%. Subsequently, we evaluated the performance of the best performing model on the second, third, and fourth cohorts (Figure 1D). When applying the CRANE pipeline on our training cohort, we used the built-in Monte Carlo k-fold cross-validation with 10 folds for training and validation, then evaluating the best performing model exactly like our SSL-attention model. All ground truth labels were available on the level of slides. All statistics were calculated on the level of slides. The primary evaluation metric was the area under the receiver operating curve (AUROC). For cross-validation, we calculated the mean performance as the mean of all AUROCs from all folds of all repetitions, together with the 95% confidence interval (95% CI) calculated by assuming a normalized distribution of AUROCs and using its standard error of the mean to identify the boundaries. For the deployment on the validation cohorts, we calculated the 95% CI of the AUROCs applying 10 000 times stratified bootstrapping with replacement. For the multiclass prediction, we used micro-averaging to obtain an overall AUROC of the experiments. We calculated P-values for each class in each experiment using a two-sided t-test and, for cross-validation, averaged these values over folds and repetitions of the experiments. The P-value in this case is a measure of distinctiveness of the classes’ prediction scores within the normalized range (zero to one) for each test set. We used the ‘metrics’ module of ‘scikit learn’ (https://scikit-learn.org/stable/modules/model_evaluation.html) to calculate accuracy, average precision, precision, recall, and F1-score. For visualization approaches, we deployed the best performing model on the test cohorts. All experiments were run on local desktop workstations with Nvidia RTX Quadro 8000 graphics processing units (GPUs).
Visualization and explainability
We plotted three tiles for the four slides of each validation cohort giving the highest bag label scores for the binarized prediction of (true) rejection when deploying the best performing model. Additionally, we generated Grad-CAM images for these tiles to get a better understanding of the model’s attention.26 To gain further insight into our model’s decisions, we generated heat maps showing the attention, as well as the attention multiplied by the prediction scores.
Code availability
All source codes for pre-processing are available at https://github.com/KatherLab/preprocessing-ng. All source codes for deep learning are available at https://github.com/KatherLab/marugoto.
Results
Deep learning can predict rejection and rejection grade from pathology images
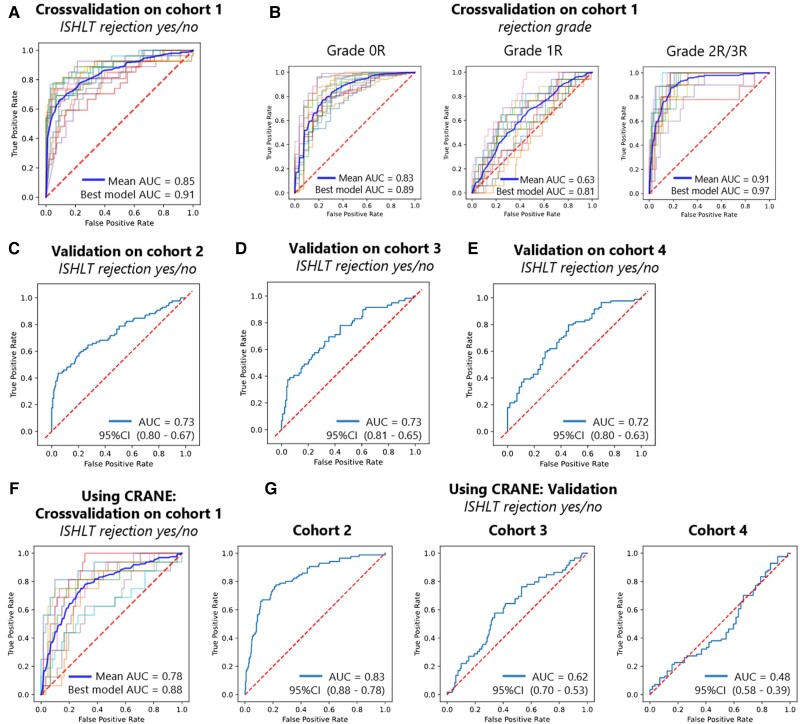
We trained an attention-based multiple instance deep learning algorithm on bags of features, extracted from patches of whole slide images. In the cross-validated experiment carried out on Cohort 1, we found a mean AUROC of 0.849 (95% CI 0.822–0.877) for binary prediction (rejection yes/no) (Figure 2A, see Supplementary material online, Table S2 for individual results). The best fold’s AUROC was 0.910 with a P-value of <0.001. For the prediction of the ISHLT grades 0R, 1R, and 2/3R, the mean AUROCs were 0.835 (95% CI 0.807–0.862), 0.633 (95% CI 0.582–0.684), and 0.905 (95% CI 0.874–0.937), respectively (Figure 2B, see Supplementary material online, Table S3 for individual results). The micro-averaged AUROC for this task was 0.814 (95% CI 0.773–0.854). The best fold’s AUROCs for this task were 0.890, 0.808, and 0.968, respectively, with a P-value <0.001 and a micro-averaged AUROC of 0.885. These results show the capacity of our network to predict rejection and rejection grade directly from histopathology images.
Figure 2.
Deep learning can predict rejection and rejection grade from pathology images. Receiver operating characteristic curves (ROC) and area under the receiver operating curve (AUC) with its 95% confidence interval (CI), as measure of performance of the classifier for heart transplant rejection following 2004 revision of the International Society for Heart and Lung Transplantation (ISHLT) grading system. Showing binarized prediction (ISHLT rejection yes/no) (A, C, D, and E) and rejection grade (ISHLT 0R, 1R, and 2/3R) (B) for cross-validation (A and B) and external validation (C, D, and E) experiments, as well as cross-validation (F) and external validation (G) for binarized prediction (ISHLT rejection yes/no) using the CRANE algorithm.
Deep learning classifiers generalize to hold-out and external patient cohorts
To further validate the performance of our network, we deployed the best performing model for each target on three validation cohorts. The validation experiments for Cohort 2 yielded an AUROC of 0.734 (95% CI 0.665–0.800, P-value <0.001) for binary prediction (rejection yes/no) (Figure 2C). For the prediction of the ISHLT grades 0R, 1R, and 2/3R, the AUROCs were 0.764 (95% CI 0.702–0.822, P-value <0.001), 0.597 (95% CI 0.521–0.667, P-value 0.099), and 0.913 (95% CI 0.869–0.955, P-value <0.001) (see Supplementary material online, Figure S2A), respectively. The micro-averaged AUROC was 0.731 (P-value 0.021). For external validation on Cohort 3, we obtained an AUROC of 0.729 (95% CI 0.647–0.805, P-value of <0.001) (Figure 2D). For the prediction of the ISHLT grades 0R, 1R, and 2/3R, the AUROCs were 0.677 (95% CI 0.587–0.760, P-value <0.001), 0.646 (95% CI 0.560–0.731, P-value 0.028), and 0.442 (95% CI 0.220–0.655, P-value 0.417), respectively (see Supplementary material online, Figure S2B). The micro-averaged AUROC was 0.659 (P-value 0.025). The external validation on Cohort 4 yielded an AUROC of 0.716 (95% CI 0.628–0.798, P-value <0.001) on the binary task (rejection yes/no) (Figure 2E). For the prediction of the ISHLT grades 0R, 1R, and 2/3R, the AUROCs were 0.722 (95% CI 0.635–0.803, P-value <0.001), 0.601 (95% CI 0.477–0.718, P-value 0.247), and 0.805 (95% CI 0.730–0.872, P-value <0.001), respectively (see Supplementary material online, Figure S2C). The micro-averaged AUROC was 0.737 (P-value 0.042). Our findings show that our models are in principle generalizable to external patient cohorts, albeit with a performance drop which is common for deep learning classifiers.17,27
Comparison of the deep learning classifier with CRANE
We compared our method to the CRANE method, the current state of the art in rejection prediction of heart transplant tissue slides.17 We re-trained CRANE on the same training cohort and evaluated it in the same cohorts as our own model. In the training cohort, the cross-validated mean AUROC of the CRANE models for the binarized target (rejection yes/no) was 0.776 (95% CI 0.717–0.835) (Figure 2F, see Supplementary material online, Table S4 for individual results), lower than the performance obtained by our attention-MIL pipeline (0.849). The best performing CRANE model yielded an AUROC of 0.882, which was again slightly lower than the performance achieved by our in-house attention-MIL pipeline (0.910). When deploying the CRANE model to our test cohorts, we received AUROCs of 0.831 (95% CI 0.778–0.879, P-value <0.001), 0.616 (95% CI 0.529–0.700, P-value 0.077), and 0.483 (95% CI 0.387–0.581, P-value 0.931) for Cohorts 2, 3, and 4, respectively (Figure 2G), overall underperforming compared to our SSL-attention model (which reached 0.734, 0.729, and 0.716, respectively). Further statistics can be seen in Supplementary material online, Table S5. In summary, our findings show that SSL-attention-MIL outperforms CRANE.
Attempt to explain attention-based predictions
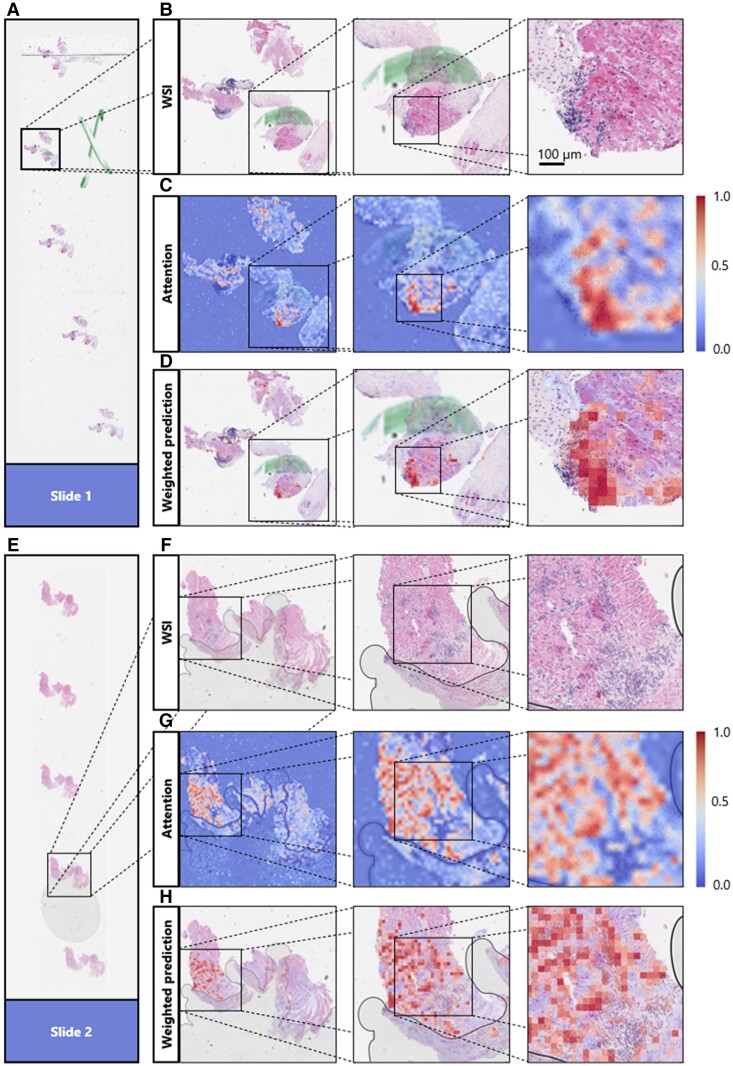
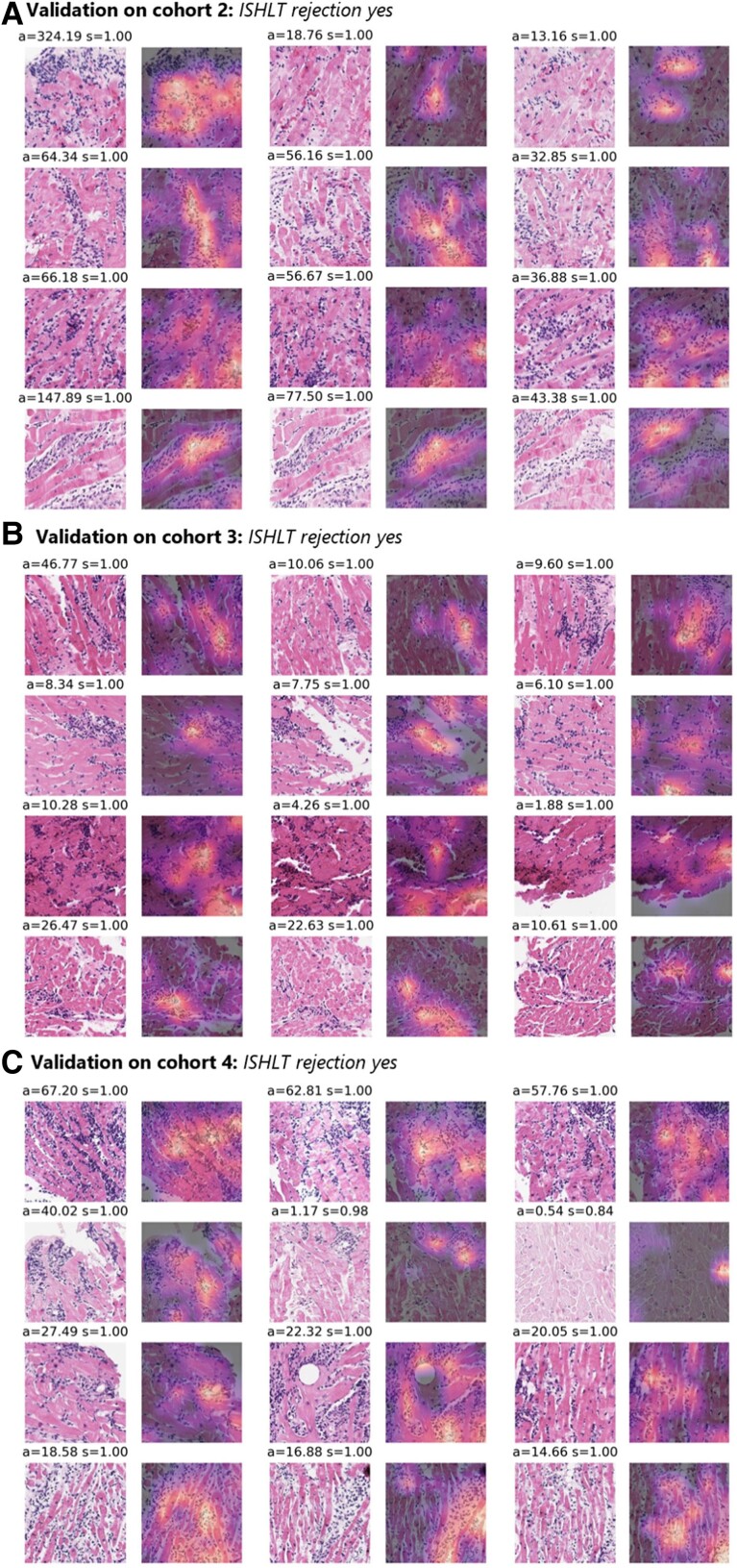
To make the model’s prediction explainable and to identify reasons for failure cases, we performed a reverse engineering task to see the spatial distribution of the network’s attention layer for the most confident true classification of binary prediction. First of all, our attention maps show that our model is concentrating only on tissue regions and not on the background or artefacts (see Figure 3C and G). This means that the presence of such artefacts (e.g. pen marks) in the test set is not problematic, and that only a simple quality control algorithm might be sufficient for clinical implementation. Analyzing whole slide attention and prediction maps on a higher resolution, we found that our model’s focus apparently seems to lie mainly in regions with a high lymphocyte density. Yet it seems to focus more on the interface of lymphocyte aggregations with the neighbouring myocardium than on these dense regions themselves (see Figure 3). We also found evidence that our model apparently was confused by the presence of a Quilty lesion,28 which was observed in a misclassified patient (see Supplementary material online, Figure S3). When analyzing the top tiles and the corresponding Grad-CAM images of the external validation cohorts, it seems that the model is concentrating on lymphocytes, confirming the findings made in heat maps at another spatial scale (Figure 4). These findings show that despite being trained on only a few hundred patients, the model has learned clinically relevant morphological patterns from whole slide images.
Figure 3.
Explaining the models’ decisions by visualizing the model’s high attention regions. Different zoom levels of areas of the whole slide images (B and F) containing one patch of the endomyocardial biopsy of two different slides (A and E) together with the attention-based heat map of the corresponding slide region (C and G) and a heat map showing the attention scores multiplied by the prediction scores (D and H). In attention-based heat maps, dark red indicates regions with a high attention, while dark blue indicates regions with a low attention [see scale in (C) and (G)]. The network is focusing on areas of the whole slide image containing tissue, ignoring artefacts, like air bubbles and pen marks (D and H). The network was trained on Cohort 1 for the binarized target (rejection yes/no) and deployed on Cohort 2. For those two slides, the network was the ‘the most confident’ about its decision (reflected by the highest attention and prediction scores). The network is highlighting regions with a high number of lymphocytes between heart muscle tissues.
Figure 4.
Explaining the models’ decisions by visualizing the model’s high attention 512 × 512 tiles. The three tiles (columns) with the highest average attention and prediction scores (attention = a, prediction score = s) for the four slides (rows) with the highest average prediction scores when deploying the best performing model to detect rejection (rejection yes/no) on the three test cohorts (A, B, and C). Together with the corresponding Grad-CAM images showing the network’s spatial attention for each of the tiles. Regions with higher attention are yellow, while regions with low attention are in dark purple. The top tiles contain many lymphocytes infiltrating the myocytes, while the network’s attention also appears to be lying on these immune cells.
Discussion
Heart transplantation remains the gold standard therapy for end-stage heart failure.29 Due to this pronounced shortage of donor organs, there is not only a need for risk adjustment tools to optimize recipient selection.30,31 In addition, a particularly good risk stratification and early adjustment of immunosuppression therapy is necessary in organ recipients because the possibility of re-transplantation is very limited. New diagnostic methods based on artificial intelligence (AI) could change and improve medical decision making in transplantation medicine in the future.18 A potential key benefit would be to reduce diagnostic uncertainty and hence reduce the need for frequent re-biopsies in the first year after transplantation, which represents a burden for healthcare systems and patients alike.
In the present study, we trained an AI method to evaluate the recognition and grading of cardiac transplantation using routine biopsies. We found high performance in the training set (by cross-validation). When deploying our model at the external validation cohorts, we found a stable, but moderate performance. A few other studies have addressed similar problems in recent years. Peyster et al.15 used handcrafted features to grade cellular rejection reporting good performance, already in 2021. Most prominently, Lipkova et al.17 presented the CRANE method, which yielded very high AUROCs in their study, after being trained on thousands of patients. Lipkova et al.17 report an external validation AUROC of around 0.83, which is better than the AUROCs of around 0.72 which we report in the validation cohorts. Yet, they report a similar decrease of performance regarding the AUROCs of around 0.12 in external validation. This is in line with other medical deep learning studies and highlights a known problem of deep learning algorithms which tend to overfit towards their training data due to their enormous number of parameters.32,33 However, our training data set comprised 10 times fewer patients, and in a head-to-head comparison of CRANE and our SSL-attention, our method outperforms CRANE. While in many computational pathology projects we have found that hyperparameter optimization does not have a pronounced impact on the final performance statistics of the whole pipeline, we admit that a more standardized approach of hyperparameter tuning might have slightly improved our performance.17,25,34,35 The small size and the underrepresentation of higher rejection gradings of the training cohort might on the other hand also be the reason for the modest generalizability of our models. It is noteworthy that we used routinely collected specimens and, contrary to Peyster et al.15, did not exclude slides from our cohorts using a quality control software. Accordingly, like in the real world, our cohorts had staining differences between slides which may impair performance. Another reason might be the presence of Quilty lesions within the ISHLT 0R group which can cause false positive predictions. Interestingly, this misclassification is also a known problem in human readers.15,36 The agreement between the CACHE-Grader model from Peyster et al. and the recorded ISHLT grade also differed between hold-out and external test set (0.89 vs. 0.83) for the binary task. Our findings are in line with other recent studies showing the usefulness of pre-training feature extractors with SSL, boosting classification performance in computational pathology.19 Our classifier also outperforms other studies which date back to the year 2017, when Tong et al.14 constructed a shallow neural network based on handcrafted features derived from 43 whole slide images (Children’s Healthcare of Atlanta cohort). This data set has been used several times afterwards, improving the performance of the cross-validated model while adopting newer methodology, but remains limited due to the very small data set size.37-39
While the application of attention heat maps is an important and fundamental step toward improved human interpretability of deep learning, they need to be interpreted with caution. This shortcoming is still seen as a major limitation for the clinical application of deep learning methodology, even though the number of Food and Drug Administration–approved tools applying deep learning in histopathology has recently been growing with speed.
Our model was only trained on cellular rejection, while graft failure is a complex process including not only additional immunological reactions like antibody-mediated rejection but also proliferative cellular processes like cardiac allograft vasculopathy. Future model development should therefore go beyond immunological rejection but try to comprise all aspects of allograft failure.
A fundamental limitation affecting all published studies is the limitation of the gold standard. The ISHLT classification itself is an imperfect predictor of clinical outcome, and future studies should train AI models directly on outcome data to overcome these limitations. This is further supported by the observation that detection and grading of heart transplant rejection can suffer from a suboptimal concordance among pathologists in the assignment of ISHLT 2004 grading of 71%, with most agreement coming from the class 0R.36 Future studies should investigate the performance of pathologists who are guided by the AI model, especially non-expert pathologists.
In summary, our study is adding evidence to existing proof-of-concept studies to show potential applications of AI systems in transplantation medicine. In particular, our study might set a new technical state of the art, which however requires validation in larger cohorts. On the other hand, our study is also a reminder that larger training cohorts of a few thousand patients are probably required for clinical-grade AI biomarkers.32,40 Future studies should compare our technical approach on larger cohorts, which could be efficiently assembled with federated or swarm learning.41,42 Our study adds to the growing evidence of AI models being to a moderate extent capable of recognizing heart transplant rejection which potentially might in the future help pathologists with pre-screening slides or standardize grading across different centres. Also, future studies should and could include multimodal input models which can in principle improve performance.43,44 Even though conclusive evidence for cost-effectiveness of AI systems in healthcare is not yet available, our study creates an incentive to investigate further development of AI in diagnostics in transplantation medicine, potentially even reducing cost and time in this sector of the healthcare system.
Supplementary material
Supplementary material is available at European Heart Journal – Digital Health.
Supplementary Material
Contributor Information
Tobias Paul Seraphin, Department of Gastroenterology, Hepatology and Infectious Diseases, University Hospital Duesseldorf, Medical Faculty at Heinrich-Heine-University, Moorenstr. 5, 40225 Dusseldorf, Germany; Department of Medicine III, University Hospital RWTH Aachen, Pauwelsstraße 30, 52074 Aachen, Germany.
Mark Luedde, Department of Cardiology and Angiology, Christian-Albrechts-University of Kiel, Arnold-Heller-Straße 3, 24105 Kiel, Germany.
Christoph Roderburg, Department of Gastroenterology, Hepatology and Infectious Diseases, University Hospital Duesseldorf, Medical Faculty at Heinrich-Heine-University, Moorenstr. 5, 40225 Dusseldorf, Germany.
Marko van Treeck, Department of Medicine III, University Hospital RWTH Aachen, Pauwelsstraße 30, 52074 Aachen, Germany.
Pascal Scheider, Institute of Pathology, RWTH Aachen University Hospital, Pauwelsstraße 30, 52074 Aachen, Germany.
Roman D Buelow, Institute of Pathology, RWTH Aachen University Hospital, Pauwelsstraße 30, 52074 Aachen, Germany.
Peter Boor, Institute of Pathology, RWTH Aachen University Hospital, Pauwelsstraße 30, 52074 Aachen, Germany.
Sven H Loosen, Department of Gastroenterology, Hepatology and Infectious Diseases, University Hospital Duesseldorf, Medical Faculty at Heinrich-Heine-University, Moorenstr. 5, 40225 Dusseldorf, Germany.
Zdenek Provaznik, Department of Cardiothoracic Surgery, University Medical Center Regensburg, Franz-Josef-Strauß-Allee 11, 93053 Regensburg, Germany.
Daniel Mendelsohn, Institute of Pathology, University of Regensburg, Franz-Josef-Strauß-Allee 11, 93053 Regensburg, Germany.
Filip Berisha, Department of Cardiology, University Heart and Vascular Center Hamburg, University Medical Center Hamburg-Hospital Eppendorf, Martinistraße 52, 20251 Hamburg, Germany; German Center for Cardiovascular Research (DZHK), Partner Site Hamburg/Kiel/Lübeck, Potsdamer Str. 58, 10785 Berlin, Germany.
Christina Magnussen, Department of Cardiology, University Heart and Vascular Center Hamburg, University Medical Center Hamburg-Hospital Eppendorf, Martinistraße 52, 20251 Hamburg, Germany; German Center for Cardiovascular Research (DZHK), Partner Site Hamburg/Kiel/Lübeck, Potsdamer Str. 58, 10785 Berlin, Germany.
Dirk Westermann, Department of Cardiology, University Heart and Vascular Center Hamburg, University Medical Center Hamburg-Hospital Eppendorf, Martinistraße 52, 20251 Hamburg, Germany; German Center for Cardiovascular Research (DZHK), Partner Site Hamburg/Kiel/Lübeck, Potsdamer Str. 58, 10785 Berlin, Germany.
Tom Luedde, Department of Gastroenterology, Hepatology and Infectious Diseases, University Hospital Duesseldorf, Medical Faculty at Heinrich-Heine-University, Moorenstr. 5, 40225 Dusseldorf, Germany.
Christoph Brochhausen, Institute of Pathology, University of Regensburg, Franz-Josef-Strauß-Allee 11, 93053 Regensburg, Germany.
Samuel Sossalla, Clinic for Cardiology and Pneumology, Georg-August University Göttingen, Robert-Koch-Straße 40, 37075 Göttingen, Germany; German Center of Cardiovascular Research (DZHK), Partner Site Göttingen, Potsdamer Str. 58, 10785 Berlin, Germany; Department of Internal Medicine II, University Medical Center Regensburg, Franz-Josef-Strauß-Allee 11, 93053 Regensburg, Germany.
Jakob Nikolas Kather, Department of Medicine III, University Hospital RWTH Aachen, Pauwelsstraße 30, 52074 Aachen, Germany; Medical Oncology, National Center for Tumor Diseases (NCT), University Hospital Heidelberg, Heidelberg, Germany; Pathology & Data Analytics, Leeds Institute of Medical Research at St James’s, University of Leeds, Leeds, United Kingdom; Else Kroener Fresenius Center for Digital Health, Technical University Dresden, Fetscherstrasse 74, 01307 Dresden, Germany.
Author contributions
T.P.S., M.L., C.R., C.B., S.S., and J.N.K. conceived the idea for the study. T.P.S. and J.N.K. performed the experiments. M.V.T. developed the software. P.S., R.D.B., P.B., Z.P., D.M., F.B., C.M., D.W., C.B., and S.S. contributed tissue samples. T.P.S. and J.N.K. wrote the initial draft of the manuscript. All authors contributed to the interpretation of the results and the editing of the final manuscript and gave approval for submission.
Funding
J.N.K. and T.L. are supported by the German Federal Ministry of Health (DEEP LIVER, ZMVI1-2520DAT111), and J.N.K. is supported by the Max-Eder-Programme of the German Cancer Aid (grant #70113864), the German Federal Ministry of Education and Research (PEARL, 01KD2104C), and the German Academic Exchange Service (SECAI, 57616814). T.L. is supported by the European Research Council (ERC; Consolidator Grant No. 771083). S.S. is funded by the German Research Foundation (DFG) through the research grant SO 1223/4-1. P.B. is supported by the German Research Foundation (DFG; Project-IDs 322900939, 454024652, and 445703531), the European Research Council (ERC; Consolidator Grant No. 101001791), the German Federal Ministry of Health (DEEP LIVER, ZMVI1-2520DAT111), and the German Federal Ministry of Economic Affairs and Energy (EMPAIA, No. 01MK2002A). M.L. is supported by the German Foundation for the chronically ill (III. Grant). C.M. is funded by the German Center for Cardiovascular Research, Deutsche Stiftung für Herzforschung, and Rolf M. Schwiete Stiftung. This research project is supported by the START-Program of the Faculty of Medicine of the RWTH Aachen University (148/21 to R.D.B.).
Data availability
The data underlying this article are not publicly available but can be requested within scientific collaboration projects from the respective data owners at the institutions.
Ethics approval
This study was carried out in accordance with the Declaration of Helsinki. This study is a retrospective analysis of digital images of anonymized archival tissue samples from three patient cohorts. Collection and anonymization of patients in all cohorts took place in each contributing centre. For the Regensburg cohorts, ethical approval was granted by the Ethical Review Board of the University Regensburg (ID: 21-2620-104). For the Hamburg and Aachen cohort, ethical approval of this retrospective investigation was not legally required due to local regulations (Berufsordnung fuer Aerzte). Nevertheless, the retrospective analysis of samples from Aachen and other collaborating centres was assessed by and approved by the Ethics Commission of the Medical Faculty of RWTH Aachen University (EK315/19), confirming that no specific patient consent is required for this retrospective study of anonymized tissue samples.
References
- 1. McDonagh TA, Metra M, Adamo M, Gardner RS, Baumbach A, Böhm M, et al. 2021 ESC guidelines for the diagnosis and treatment of acute and chronic heart failure. Eur Heart J 2021;42:3599–3726. [DOI] [PubMed] [Google Scholar]
- 2. Lund LH, Khush KK, Cherikh WS, Goldfarb S, Kucheryavaya AY, Levvey BJ, et al. The Registry of the International Society for Heart and Lung Transplantation: thirty-fourth adult heart transplantation report-2017; focus theme: allograft ischemic time. J Heart Lung Transplant 2017;36:1037–1046. [DOI] [PubMed] [Google Scholar]
- 3. Ruiz-Ortiz M, Rodriguez-Diego S, Delgado M, Kim J, Weinsaft JW, Ortega R, et al. Myocardial deformation and acute cellular rejection after heart transplantation: impact of inter-vendor variability in diagnostic effectiveness. Echocardiography 2019;36:2185–2194. [DOI] [PubMed] [Google Scholar]
- 4. van Heeswijk RB, Piccini D, Tozzi P, Rotman S, Meyer P, Schwitter J, et al. Three-dimensional self-navigated T2 mapping for the detection of acute cellular rejection after orthotopic heart transplantation. Transplant Direct 2017;3:e149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Stewart S, Winters GL, Fishbein MC, Tazelaar HD, Kobashigawa J, Abrams J, et al. Revision of the 1990 working formulation for the standardization of nomenclature in the diagnosis of heart rejection. J Heart Lung Transplant 2005;24:1710–1720. [DOI] [PubMed] [Google Scholar]
- 6. Angelini A, Andersen CB, Bartoloni G, Black F, Bishop P, Doran H, et al. A web-based pilot study of inter-pathologist reproducibility using the ISHLT 2004 working formulation for biopsy diagnosis of cardiac allograft rejection: the European experience. J Heart Lung Transplant 2011;30:1214–1220. [DOI] [PubMed] [Google Scholar]
- 7. Tizhoosh HR, Diamandis P, Campbell CJV, Safarpoor A, Kalra S, Maleki D, et al. Searching images for consensus: can AI remove observer variability in pathology? Am J Pathol 2021;191:1702–1708. [DOI] [PubMed] [Google Scholar]
- 8. Echle A, Rindtorff NT, Brinker TJ, Luedde T, Pearson AT, Kather JN. Deep learning in cancer pathology: a new generation of clinical biomarkers. Br J Cancer 2020;124:686–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cifci D, Foersch S, Kather JN. Artificial intelligence to identify genetic alterations in conventional histopathology. J Pathol 2022;257:430–444. [DOI] [PubMed] [Google Scholar]
- 10. Ilse M, Tomczak JM, Welling M. Attention-based deep multiple instance learning. In: Dy J, Krause A, ed. Proceedings of the 35th International Conference on Machine Learning. PMLR; 2018, p2127–2136.
- 11. Shmatko A, Ghaffari Laleh N, Gerstung M, Kather JN. Artificial intelligence in histopathology: enhancing cancer research and clinical oncology. Nat Cancer 2022;3:1026–1038. [DOI] [PubMed] [Google Scholar]
- 12. Ghaffari Laleh N, Muti HS, Loeffler CML, Echle A, Saldanha OL, Mahmood F, et al. Benchmarking weakly-supervised deep learning pipelines for whole slide classification in computational pathology. Med Image Anal 2022;79:102474. [DOI] [PubMed] [Google Scholar]
- 13. Kers J, Bülow RD, Klinkhammer BM, Breimer GE, Fontana F, Abiola AA, et al. Deep learning-based classification of kidney transplant pathology: a retrospective, multicentre, proof-of-concept study. Lancet Digit Health 2022;4:e18–e26. [DOI] [PubMed] [Google Scholar]
- 14. Tong L, Hoffman R, Deshpande SR, Wang MD. Predicting heart rejection using histopathological whole-slide imaging and deep neural network with dropout. In: 2017 IEEE EMBS International Conference on Biomedical Health Informatics (BHI), Piscataway, NJ. IEEE EMBS; 2017, p1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Peyster EG, Arabyarmohammadi S, Janowczyk A, Azarianpour-Esfahani S, Sekulic M, Cassol C, et al. An automated computational image analysis pipeline for histological grading of cardiac allograft rejection. Eur Heart J 2021;42:2356–2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Laleh NG, Ligero M, Perez-Lopez R, Kather JN. Facts and hopes on the use of artificial intelligence for predictive immunotherapy biomarkers in cancer. Clin Cancer Res 2022;29:316–323. [DOI] [PubMed] [Google Scholar]
- 17. Lipkova J, Chen TY, Lu MY, Chen RJ, Shady M, Williams M, et al. Deep learning-enabled assessment of cardiac allograft rejection from endomyocardial biopsies. Nat Med 2022;28:575–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mahmood F, Topol EJ. Digitising heart transplant rejection. Lancet 2022;400:17. [DOI] [PubMed] [Google Scholar]
- 19. Schirris Y, Gavves E, Nederlof I, Horlings HM, Teuwen J. DeepSMILE: contrastive self-supervised pre-training benefits MSI and HRD classification directly from H&E whole-slide images in colorectal and breast cancer. Med Image Anal 2022;79:102464. [DOI] [PubMed] [Google Scholar]
- 20. Bossuyt PM, Reitsma JB, Bruns DE, Gatsonis CA, Glasziou PP, Irwig L, et al. STARD 2015: an updated list of essential items for reporting diagnostic accuracy studies. BMJ 2015;351:h5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Muti HS, Loeffler C, Echle A, Heij LR, Buelow RD, Krause J, et al. The Aachen protocol for deep learning histopathology: a hands-on guide for data preprocessing. Zenodo 2020. [Google Scholar]
- 22. Saldanha OL, Loeffler CML, Niehues JM, van Treeck M, Seraphin TP, Hewitt KJ, et al. Self-supervised deep learning for pan-cancer mutation prediction from histopathology. bioRxiv 2022. doi: 10.1101/2022.09.15.507455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang X, Du Y, Yang S, Zhang J, Wang M, Zhang J, et al. RetCCL: clustering-guided contrastive learning for whole-slide image retrieval. Med Image Anal 2022;83:102645. [DOI] [PubMed] [Google Scholar]
- 24. Kingma DP, Ba J. Adam: a method for stochastic optimization. arXiv 2014;1412.6980:v9. [Google Scholar]
- 25. Lu MY, Williamson DFK, Chen TY, Chen RJ, Barbieri M, Mahmood F. Data-efficient and weakly supervised computational pathology on whole-slide images. Nat Biomed Eng 2021;5:555–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Selvaraju RR, Cogswell M, Das A. Grad-cam: visual explanations from deep networks via gradient-based localization. Proc Estonian Acad Sci Biol Ecol 2017;1:618–626. [Google Scholar]
- 27. Echle A, Ghaffari Laleh N, Quirke P, Grabsch HI, Muti HS, Saldanha OL, et al. Artificial intelligence for detection of microsatellite instability in colorectal cancer-a multicentric analysis of a pre-screening tool for clinical application. ESMO Open 2022;7:100400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Forbes RD, Rowan RA, Billingham ME. Endocardial infiltrates in human heart transplants: a serial biopsy analysis comparing four immunosuppression protocols. Hum Pathol 1990;21:850–855. [DOI] [PubMed] [Google Scholar]
- 29. McDonagh TA, Metra M, Adamo M. 2021 ESC guidelines for the diagnosis and treatment of acute and chronic heart failure: developed by the task force for the diagnosis and treatment of acute and chronic heart failure of the European Society of Cardiology (ESC). With the special contribution of the Heart Failure Association (HFA) of the ESC. Eur Heart J 2021;42:3599–3726.34447992 [Google Scholar]
- 30. Schramm R, Zittermann A, Fuchs U, Fleischhauer J, Costard-Jäckle A, Ruiz-Cano M, et al. Donor-recipient risk assessment tools in heart transplant recipients: the Bad Oeynhausen experience. ESC Heart Fail 2021;8:4843–4851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sunavsky J, Fujita B, Ensminger S, Börgermann J, Morshuis M, Fuchs U, et al. Predictors of failure after high urgent listing for a heart transplant. Interact Cardiovasc Thorac Surg 2018;27:950–957. [DOI] [PubMed] [Google Scholar]
- 32. Echle A, Grabsch HI, Quirke P, van den Brandt PA, West NP, Hutchins GGA, et al. Clinical-grade detection of microsatellite instability in colorectal tumors by deep learning. Gastroenterology 2020;159:1406–1416.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. McKinney SM, Sieniek M, Godbole V, Godwin J, Antropova N, Ashrafian H, et al. International evaluation of an AI system for breast cancer screening. Nature 2020;577:89–94. [DOI] [PubMed] [Google Scholar]
- 34. Kather JN, Pearson AT, Halama N, Jäger D, Krause J, Loosen SH, et al. Deep learning can predict microsatellite instability directly from histology in gastrointestinal cancer. Nat Med 2019;25:1054–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kather JN, Heij LR, Grabsch HI, Loeffler C, Echle A, Muti HS, et al. Pan-cancer image-based detection of clinically actionable genetic alterations. Nat Cancer 2020;1:789–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Crespo-Leiro MG, Zuckermann A, Bara C, Mohacsi P, Schulz U, Boyle A, et al. Concordance among pathologists in the second Cardiac Allograft Rejection Gene Expression Observational Study (CARGO II). Transplantation 2012;94:1172–1177. [DOI] [PubMed] [Google Scholar]
- 37. Dooley AE, Tong L, Deshpande SR, Wang MD. Prediction of heart transplant rejection using histopathological whole-slide imaging. IEEE EMBS Int Conf Biomed Health Inform 2018;1:251–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhu Y, Wang MD, Tong L, Deshpande SR. Improved prediction on heart transplant rejection using convolutional autoencoder and multiple instance learning on whole-slide imaging. IEEE EMBS Int Conf Biomed Health Inform 2019;1:1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Giuste F, Venkatesan M, Zhao C, Tong L, Zhu Y, Deshpande SR, et al. Automated classification of acute rejection from endomyocardial biopsies . In: Proceedings of the 11th ACM International Conference on Bioinformatics, Computational Biology and Health Informatics. New York, NY, USA: Association for Computing Machinery; 2020, p1–9. [Google Scholar]
- 40. Campanella G, Hanna MG, Geneslaw L, Miraflor A, Werneck Krauss Silva V, Busam KJ, et al. Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nat Med 2019;25:1301–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lu MY, Chen RJ, Kong D, Lipkova J, Singh R, Williamson DFK, et al. Federated learning for computational pathology on gigapixel whole slide images. Med Image Anal 2022;76:102298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Saldanha OL, Quirke P, West NP, James JA, Loughrey MB, Grabsch HI, et al. Swarm learning for decentralized artificial intelligence in cancer histopathology. Nat Med 2021;28:1232–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lipkova J, Chen RJ, Chen B, Lu MY, Barbieri M, Shao D, et al. Artificial intelligence for multimodal data integration in oncology. Cancer Cell 2022;40:1095–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Howard FM, Kather JN, Pearson AT. Multimodal deep learning: an improvement in prognostication or a reflection of batch effect? Cancer Cell 2022;41:5–6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are not publicly available but can be requested within scientific collaboration projects from the respective data owners at the institutions.