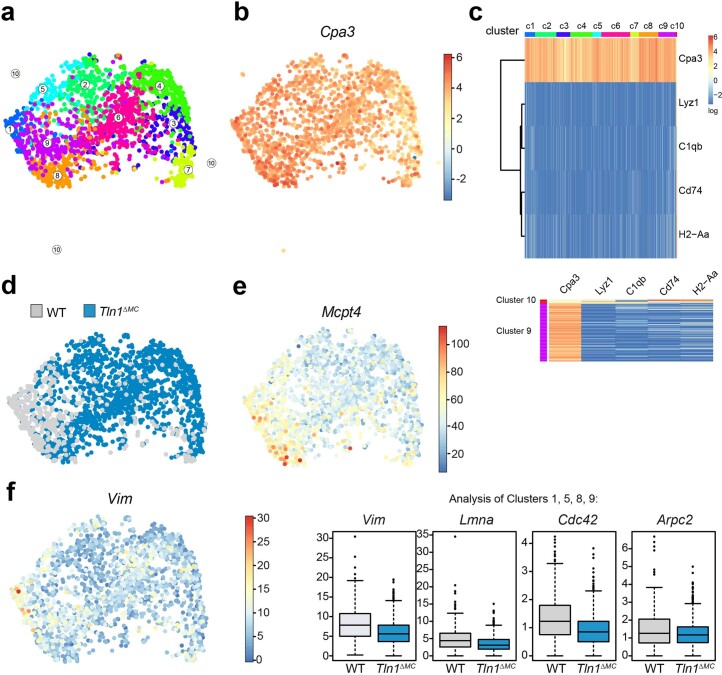
Extended Data Fig. 7. Single cell RNAseq analysis of dermal MCs.
(a) UMAP of single-cell transcriptomes of sorted CD45+Lin−YFP+ cells (WT and Tln1∆MC MCs combined) highlighting RaceID3 clusters, displaying clusters 1–10. The outlying cluster 10 was not displayed in the corresponding Fig. 4b, as it carried an additional macrophage gene signature (see Extended Data Fig. 7c). Numbers denote clusters. n = 1895 cells derived from three WT and three Tln1∆MC mice). (b) UMAP showing the expression of the gene encoding the MC marker CPA3 in clusters 1–10. The color bar indicates log2 normalized expression. (c) Heatmap showing Cpa3 expression and genes specifically expressed in cluster 10. The color bar at the top of the heatmap indicates the RaceID3 clusters. Scale bar, log2 normalized expression. Below, the heatmap of clusters 10 and 9 are also displayed at higher magnification. (d) UMAP of single-cell transcriptomes of MCs highlighting the sources or origins of the cells, that is which mice (WT or Tln1∆MC) the cells were isolated from. This is a duplicate of Fig. 4c. (e) UMAP of Mcpt4 expression. The color bar indicates normalized transcript counts. (f) UMAP of Vim expression (left). The color bar indicates normalized transcript counts. Box plots show analysis of clusters 1, 5, 8, 9 and the expression of genes encoding the cytoskeletal elements VIM, LMNA, CDC42 and ARPC2 in WT and Tln1∆MC cells (right). The expression of the genes was significantly downregulated in MCs from Tln1∆MC relative to WT mice. Boxes indicate 1st and 3rd quartiles and the black line indicates the median. Whiskers indicate 5% and 95% quantiles. For all comparisons: Benjamini-Hochberg corrected P < 0.05. Vim: P = 4.1e-20, Lmna: P = 9.3e-18, Cdc42: P = 1e-9, Arpc2: P = 0.049.