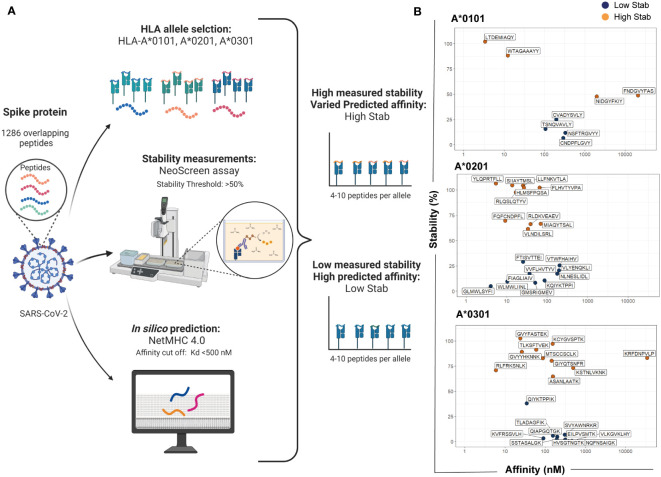
Figure 1.
Selection of SARS-CoV-2 spike peptides based on stability measurements and affinity predictions. Workflow for selecting and grouping peptides for the experiment design. (A) An overlapping 9-mer peptide library from the spike protein (total of 1286 peptides) was analyzed for stability using NeoScreen® assay and the same peptides were analyzed with NetMHC4.0 for affinity predictions. Assays and predictions were done in the context of HLA-A*0101, -A*0201 and -A*0301. The figure was created in BioRender. (B) For each allele, peptides were divided in two groups named High Stab for peptides with high measured stability (>50%) and varied predicted affinity (orange dots) or Low Stab for peptides with low measured stability (<50%) and a predicted affinity for binders (<500nM) (blue dots).