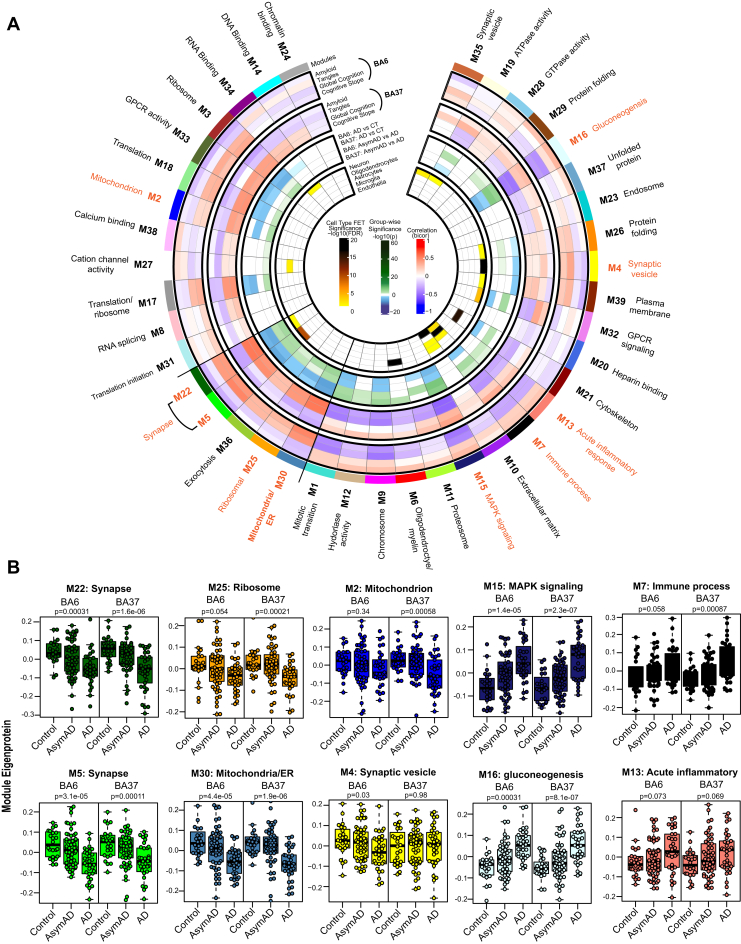
Fig. 2.
Consensus correlation network of a multi-region human brain proteome.A, a consensus correlation network (cWGCNA) was constructed with 7787 proteins across BA6 and BA37 and yielded 39 co-expression protein modules. In the inner most heatmap, enrichment of cell-type markers (as determined by one-way Fisher’s exact test) for each module is visualized for neuronal, oligodendrocyte, astrocyte, microglial, and endothelial cell types. The panel outside of the cell type results highlights group-wise differences in module eigenproteins for AD versus Control and AsymAD versus AD in each brain region. The two outer most heatmaps depict the correlation (Bicor) of module eigenproteins with pathological (Amyloid and Tangle burden) and clinical (Global cognitive function and cognitive slope) phenotypes for both brain regions. Modules are identified by color and number, accompanied by top gene ontology (GO) terms representative of modular biology. Scale bars for cell-type enrichment (darker color indicates stronger enrichment), weighted group-wise eigenprotein difference (darker green correspond to a stronger positive relationship and deeper blue indicates a stronger negative relationship), and bidirectional module—trait relationships (red indicating positive correlation and blue indicating negative correlation) are at the center of the plot. B, module eigenproteins (MEs) grouped by diagnosis (Control, AsymAD, and AD) were plotted as box and whisker plots for modules of interest, chosen based on their preservation in AsymAD compared to AD and relationships to cognitive measures. MEs were compared in each brain region using one-way ANOVA, unadjusted p values are shown. Box plots represent median, 25th, and 75th percentiles. Box hinges represent the interquartile range of the two middle quartiles with a group. Error bars are based on data points 1.5 times the interquartile range from the box hinge. AD, Alzheimer’s disease; AysmAD, asymptomatic AD; cWGCNA, consensus weighted gene correlation network analysis; BA6, Brodmann area 6; BA37, Brodmann area 37.