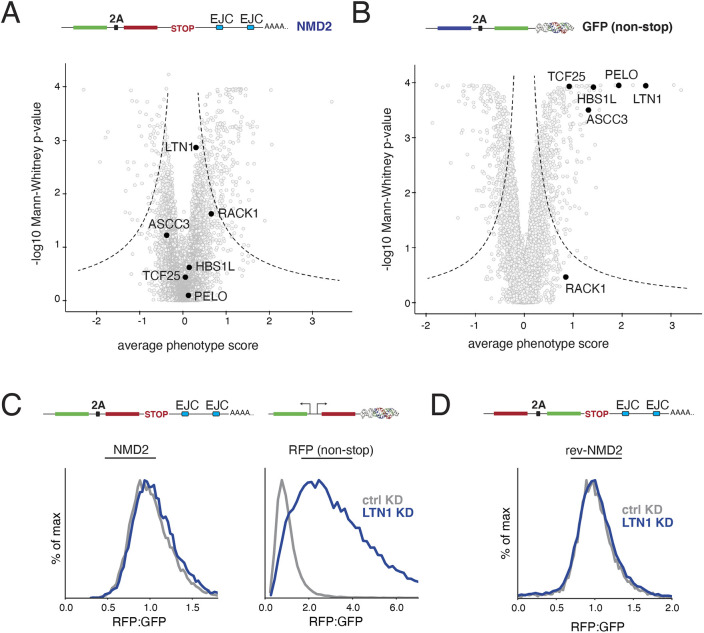
Fig. 4.
NMD-linked protein degradation is not mediated by the canonical RQC pathway. (A) Volcano plot of the NMD2 reporter CRISPRi screen as in Fig. 3A. Highlighted in black are factors involved in the canonical RQC (LTN1, RACK1, ASCC3, HBS1L, TCF25 and PELO). (B) For comparison, RQC factors are highlighted in black on a volcano plot for an earlier CRISPRi screen using a non-stop reporter (consisting of a BFP, a viral 2A skipping sequence and a GFP conjugated a triple helix moiety to stabilize the mRNA transcript, which would usually be degraded due to the lack of a stop codon) conducted using identical conditions as in A (Hickey et al., 2020). (C) K562 CRISPRi cells stably expressing either an inducible NMD2 reporter or a constitutively expressed non-stop reporter with matched GFP and RFP fluorophores (in this case, a functionally equivalent non-stop reporter with two separate promoters, one driving GFP, and the other RFP conjugated to the triple helix moiety; as in Hickey et al., 2020) were infected with a sgRNA targeting the E3 ligase LTN1. The RFP to GFP ratios for NMD2, and the GFP to RFP ratio for the non-stop reporter as determined by flow cytometry are displayed as a histogram and are quantified in Fig. S4B. (D) K562 CRISPRi cells expressing a reversed version of the NMD2 reporter (rev-NMD2) were infected with an sgRNA against LTN1 and analyzed as in C and are quantified in Fig. S4C.