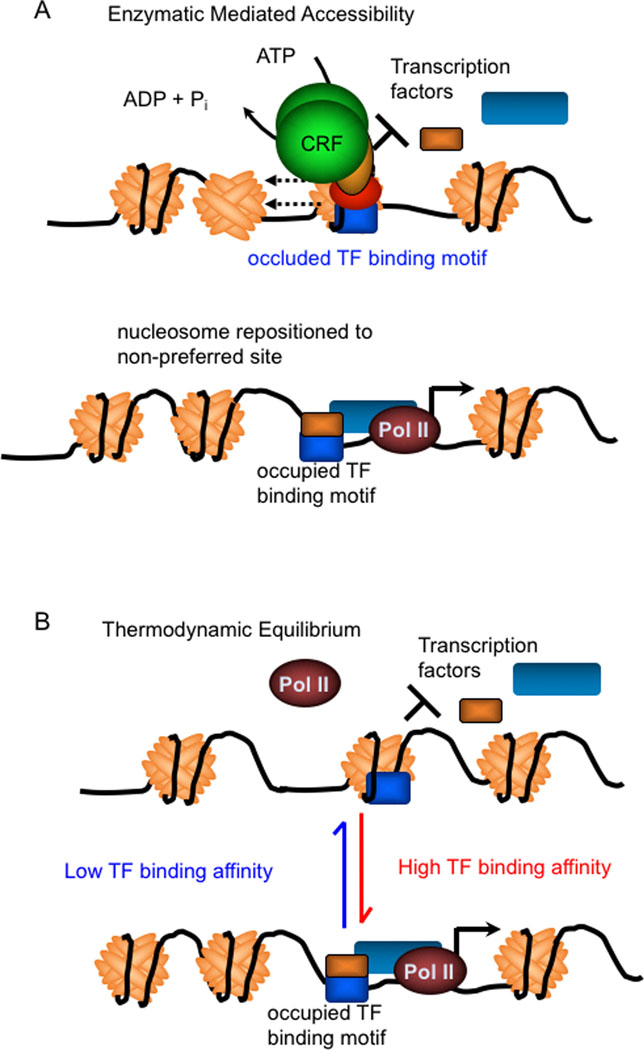
Fig. 2. Nucleosome organization in the genome is established by chromatin remodelling machines and competition with other DNA-binding regulatory factors.
(A) Nucleosome organization, especially near TSSs and other cis-regulatory regions is constrained by chromatin remodelling factor (CRF) enzymes that use the energy released from hydrolysis of ATP to catalyze repositioning of nucleosomes on DNA. Diverse assemblies of chromatin remodelling complexes built around one of four families of ATPases provide specificity. However, most subunits with these chromatin remodelling complexes lack DNA-binding domains that provide sequence-specificity, and require conventional sequence specific TFs to guide CRFs to their appropriate targets. (B) The histones in nucleosomes bind to DNA with varying affinities, and nucleosomes prefer to occupy certain sequences more than others. Competition between nucleosomes and other DNA-binding factors influences the positioning and occupancy of nucleosomes on the genome. TF-directed recruitment of ATPase-dependent remodellers to specific genes probably facilitates formation of nucleosomes on non-preferred sequences instead of regulatory sequences actively bound by their cognate TFs.