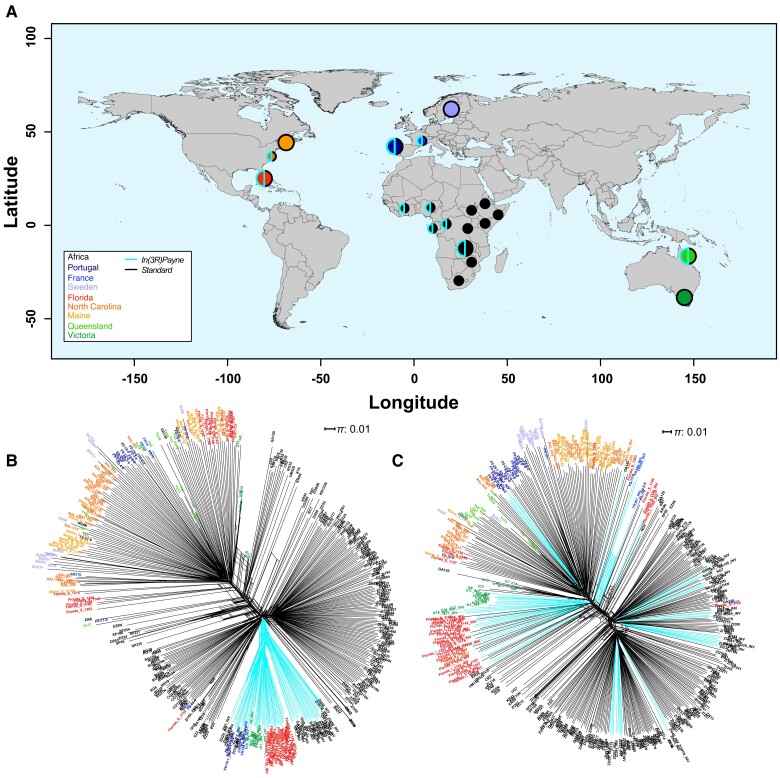
Fig. 1.
Distribution of samples and phylogenetic relationships among In(3R)Payne karyotypes across four continents. (A) Geographic origin of the samples used in this study. The color code indicates the continent where flies were sampled (Africa, Europe, North America, and Australia). The outline of the circles indicates whether the samples contain chromosomes with In(3R)Payne (in cyan) and/or with the standard arrangement (in black); the size of the circles indicates whether samples were used only for phylogenetic reconstruction (small circles) or in addition also for karyotype-specific genomic analyses (large circles). (B) Haplotype network constructed from 3766 SNPs within the breakpoints of In(3R)Payne; cyan edges represent samples with In(3R)Payne, whereas black edges represent samples with the standard arrangement. (C) Haplotype network based on 4,849 randomly drawn SNPs at a distance of >200 kb from In(3L)P and In(3R)Payne (see Materials and Methods). See table 1 for statistical analyses. Note that several haplotypes from Florida cluster with the NG9 reference strain (see fig. 1B and C). This may be an artifact of our bioinformatic method for haplotype reconstruction (see Materials and Methods); we therefore excluded these samples from downstream analyses.