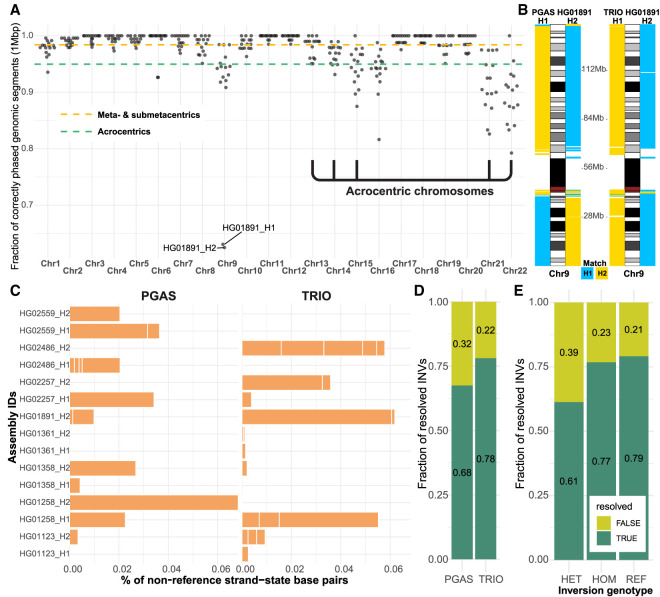
Figure 2.
Phasing accuracy and inversion analysis of trio-based and trio-free assemblies. (A) Phasing accuracy of PGAS (trio-free) assemblies with respect to trio-based phasing. (B) Haplotype assignment of 1-Mbp-sized blocks (left from ideogram, H1; right from ideogram, H2) to either haplotype 1 or 2 (blue, H1; yellow, H2) using single-nucleotide polymorphisms phased using trio information (1000 Genomes Project panel) with respect to the reference (GRCh38). (C) A barplot reporting the percentage of base pairs in an opposite (reverse) orientation in contrast to the expected (direct) orientation based on Strand-seq analysis of assembly directionality, shown separately for trio-free (PGAS, n = 15; left) and trio-based (TRIO, n = 23; right) assemblies. (D) Fraction of tested inversion sites that are fully informative (TRUE; dark green). (E) Fraction of tested inversion sites that are fully informative (TRUE; dark green) as a function of inversion genotype. (HET) Heterozygous, (HOM) homozygous inverted, (REF) homozygous reference.