Figure 2.
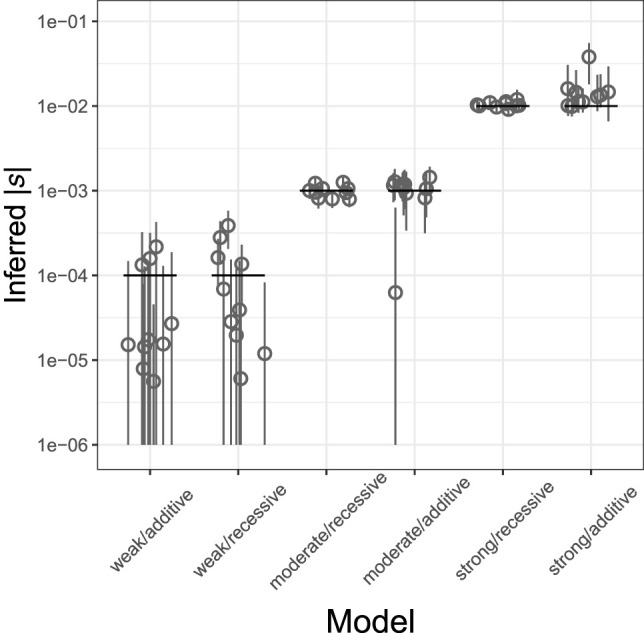
Inference of s from a genome-wide set of deleterious SNPs for different strengths of selection (weak: s = −0.0001; moderate: s = −0.001; strong: s = −0.01) and dominance effects (recessive: h = 0; additive: h = 0.5). Each scenario includes the estimates from 10 simulated data sets. True values are shown as black horizontal segments, with medians of the inferred posterior distributions denoted by gray circles and their 95% credible intervals by gray vertical lines. The y-axis is in log10 scale; all values are in absolute numbers. Here, 50,000 trios are used.
