Figure 7.
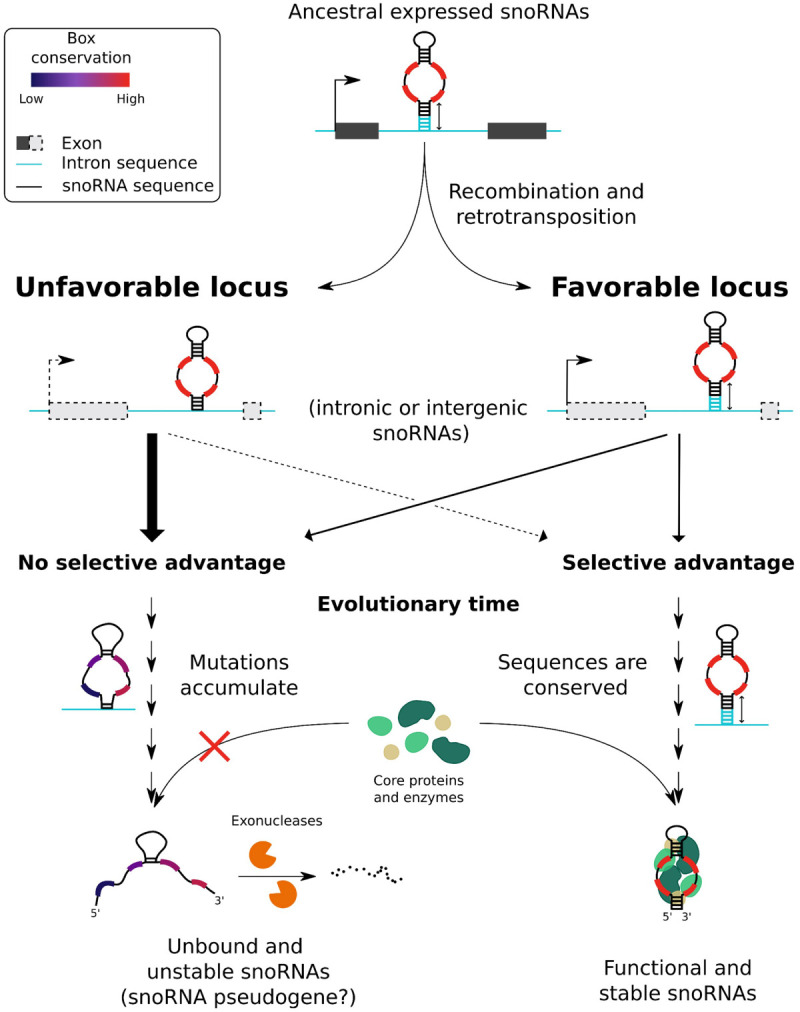
Model explaining the low proportion of expressed snoRNAs annotated in vertebrate genomes. Expressed ancestral snoRNAs presumably had box motifs close to their current consensus sequence, stable terminal stem, and global structure, as well as a genomic context favorable to their expression owing to a nearby promoter. Following a recombination or retrotransposition event, the new locus of these copied snoRNAs might prove to be favorable or unfavorable to their expression, such as containing or not a nearby promoter and flanking sequences likely to promote the formation of a stable terminal stem (the dashed line for the promoter representing a possible lack of promoter). If the newly copied snoRNA induced a gain in fitness, there likely was selective pressure to conserve its sequence and its flanking regions, promoting the binding of core proteins and enzymes to the expressed snoRNA to stabilize its structure and generate a stable and functional snoRNA. Conversely, snoRNAs integrated in unfavorable loci, if transcribed, likely had their binding to core proteins and enzymes hindered by the lack of stable terminal stem, therefore not providing a selective advantage for the organism and thereby allowing mutation accumulation within these snoRNAs. If transcribed, these unstable snoRNAs were then likely degraded by exonucleases and thus represent the nonexpressed snoRNAs (possibly snoRNA pseudogenes) in the present vertebrate genome annotations. Of note, only C/D box snoRNAs are represented in the model, but the same conclusions apply to H/ACA box snoRNAs according to our analyses.
