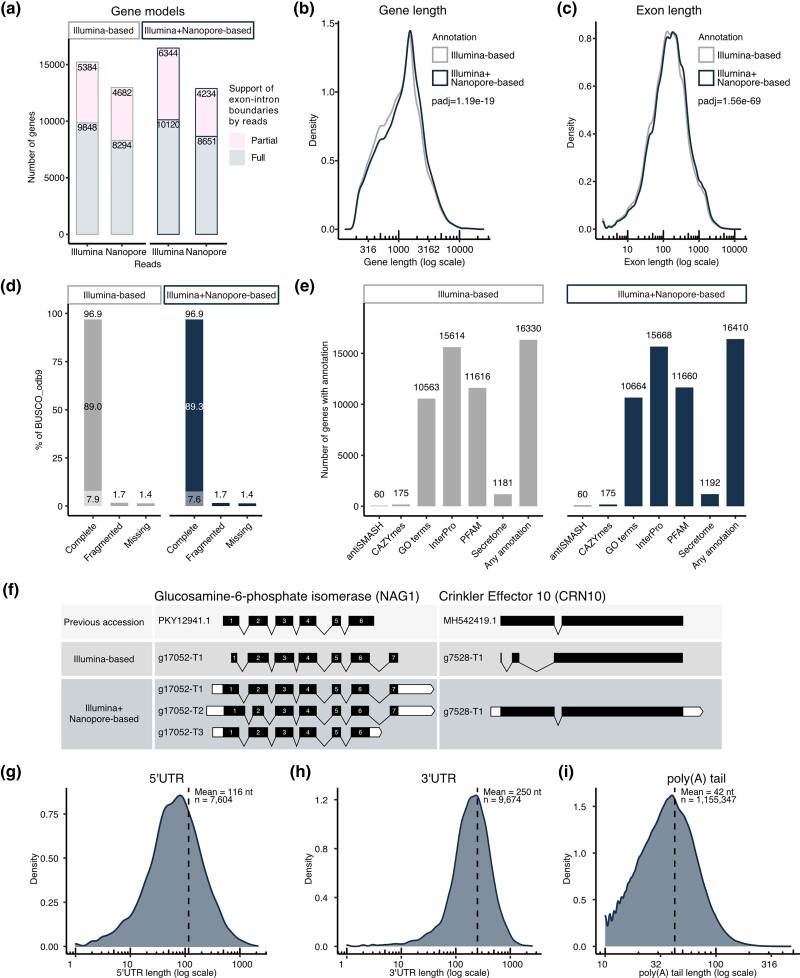
Fig. 3.
General features of revised gene models. a) Support of exon–intron boundaries of Illumina-based and Illumina + Nanopore-based gene annotations by Illumina and Nanopore RNA-Seq reads. The number of genes with all boundaries (full support) and partial boundaries (partial support) supported by experimental evidence is indicated. Exon-less genes are not displayed. Comparison of gene b) and exon c) length distribution between Illumina and Illumina + Nanopore-based gene annotations. The x-axes are on a log scale and a paired t-test was used to assess statistical significance. d) Comparison of BUSCO gene categories. The complete stack is split into single copy (top) and duplicated (bottom). e) Comparison of functional annotation of Illumina-based and Illumina + Nanopore-based gene models. f) Comparison of gene models revised using long-read data to previous annotations and Illumina-based gene models. Black boxes represent exons, lines are introns, and white boxes are UTRs. Length distribution of 5′UTRs g) and 3′UTRs h) of the Illumina + Nanopore gene models. i) Length distribution of poly(A) tails detected in spores. The x-axes are on a log scale.