Fig. 3. A subset of hepatocytes from mouse and human NASH livers exhibit enhanced EphB2 activity.
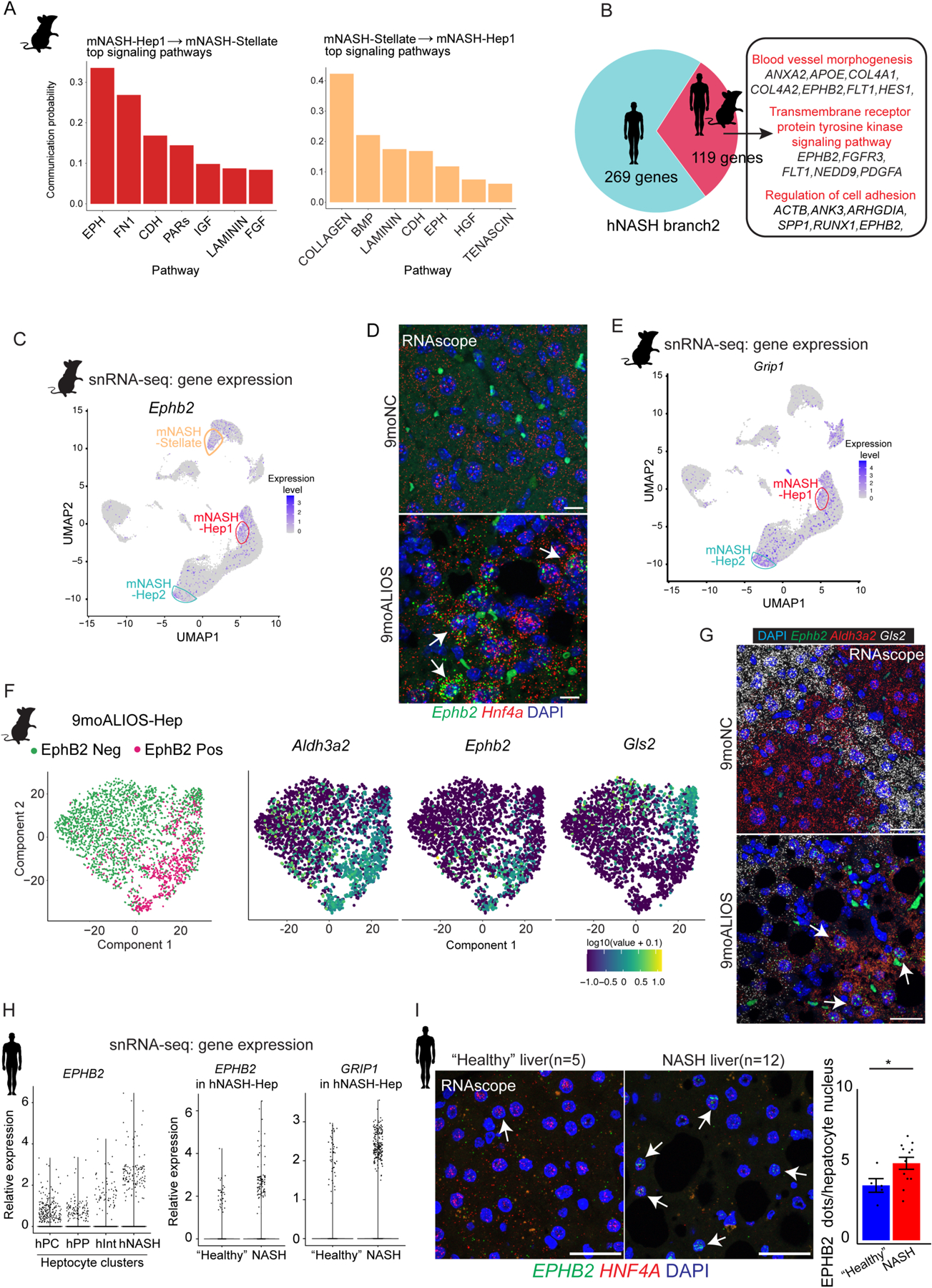
(A) Top signaling pathways mediated intercellular communication between mNASH-Hep1 and mNASH-Stellate.
(B) 119 common genes including EPHB2 between mouse and human were identified as contributing to mNASH branch/hNASH branch2.
(C) UMAP plot of snRNA-seq showing mNASH-Hep1/2 Ephb2 RNA expression.
(D) Elevated transcript abundance of Ephb2 in hepatocytes from 9moALIOS liver was validated by RNAscope (white arrows pointing to the hepatocyte nuclei labelled by Hnf4a, scale bar=20μm, n=3 in each group).
(E) UMAP plot of snRNA-seq shown Grip1 was prominently elevated in mNASH-Hep1/2.
(F) Subclustering of hepatocytes from 9moALIOS liver based on Ephb2 transcript abundance and expression pattern of zonation landmarks Aldh3a2 and Gls2 in EphB2-positive and EphB2-negative clusters.
(G) Co-staining Ephb2 with zonation landmarks using RNAscope validated that Ephb2-expressing hepatocytes mostly localized in pericentral zone (scale bar=25μm, n=3 in each group). (H) EPHB2 expression across different hepatocyte clusters in human snRNA-seq and differential expression of EPHB2 and GRIP1 between “healthy” and NASH conditions within the hNASH-Hep cluster.
(I) RNA in situ hybridization using RNAscope showed increased EPHB2 expression (green, white arrows) in hepatocyte nuclei stained with HNF4A (red) in livers from human patients with NASH (n=12) compared with “healthy” donor livers (n=5) (*p<0.05, Mann-Whitney U test, scale bar=25μm).
