Fig. 5. Hepatocyte-specific EphB2 activation in mice fed NC diet induces cell-autonomous inflammation.
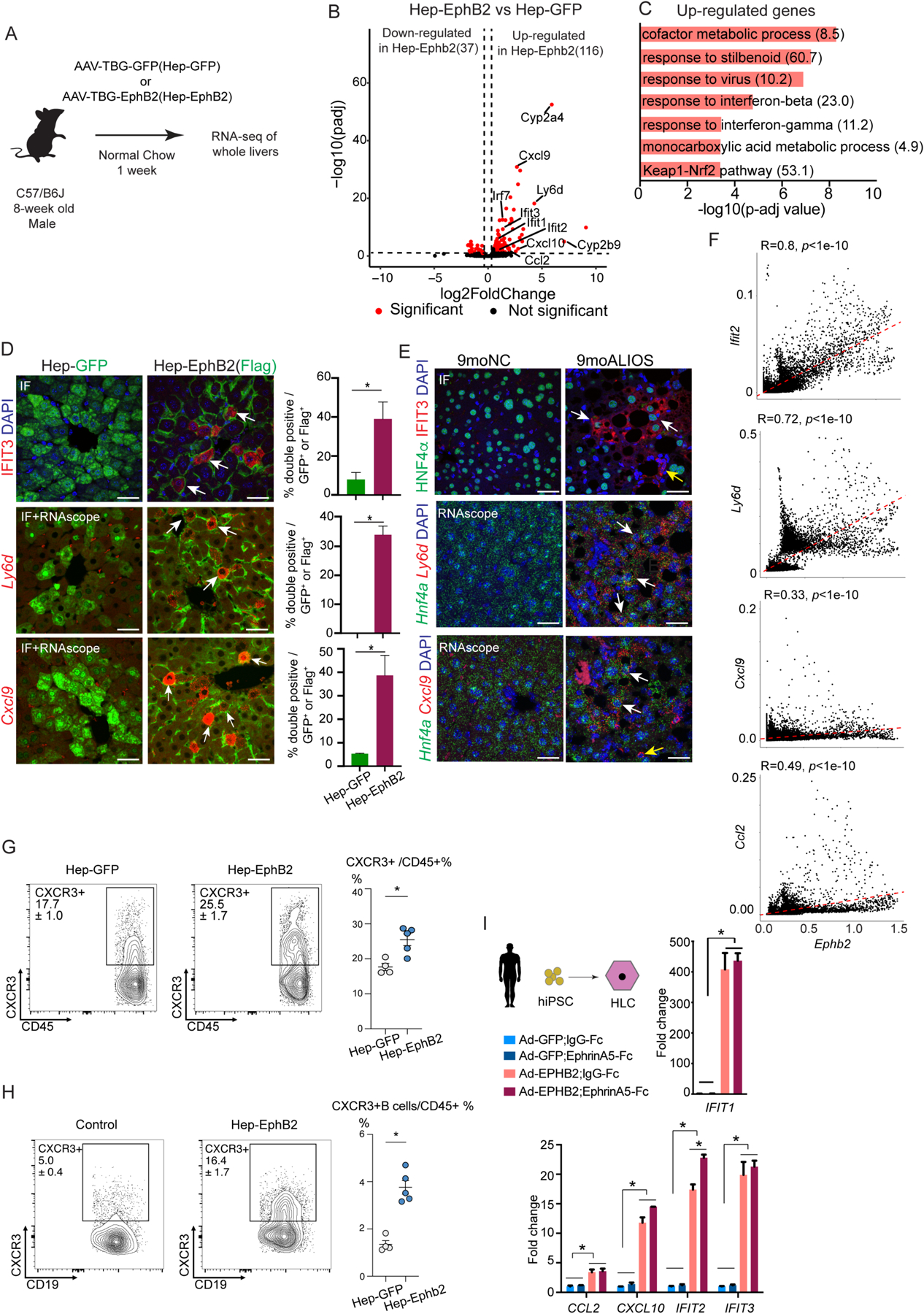
(A) Experimental scheme of RNA-seq of whole livers from the mice with forced expression of EphB2 (Hep-EphB2, n=3) or GFP (Hep-GFP, n=3).
(B) Volcano plot of the differentially expressed genes (shown as red dots with cutoff padj<0.1 and abs(log2FC) > 0.5) between Hep-EphB2 and Hep-GFP (Ephb2 is not shown on this plot).
(C) GO analysis demonstrated enrichment of metabolic processes, interferon response pathways, oxidative stress pathways, and Keap1-Nrf2 in up-regulated genes (Odds ratio indicated after each term).
(D) IF staining (IFIT3) and RNA-scope (Ly6d and Cxcl9) showing induced inflammatory response in hepatocytes with ectopic expression of EphB2 (white arrows). GFP and Flag antibodies stained the transduced hepatocytes from AAV-GFP and AAV-EphB2-Flag tail-vein injected mouse livers respectively. Data are expressed as mean ± SD, n=3 in each group, *p<0.05, Mann-Whitney U test, scale bar=25μm.
(E) IF staining (IFIT3) and RNA-scope (Ly6d and Cxcl9) in hepatocytes from mice treated with ALIOS diet for 9 months (white arrows). Antibody against Hnf4α or RNAscope probe detecting Hnf4a was used to label hepatocytes. Ifit3 and Cxcl9 were also detected in the non-hepatocytes in 9moALIOS livers (yellow arrows, scale bar=25μm).
(F) Correlation analysis of snRNA-seq demonstrated Ifit2, Ly6d, Cxcl9, and Ccl2 positively correlated with Ephb2 expression in hepatocytes.
(G-H) Flow cytometry showing an increased percentage of CXCR3 and CXCR3+ B cells in Hep-Ephb2 livers. Data are expressed as mean ± SEM, n=4/5 in each group, *p<0.05.
(I) Activation of EPHB2 in hiPSC-HLC induced inflammatory response and the effect was largely EphrinA5-independent. Data are expressed as mean ± SD, n=3 in each group, * p<0.05, Mann–Whitney U test.
