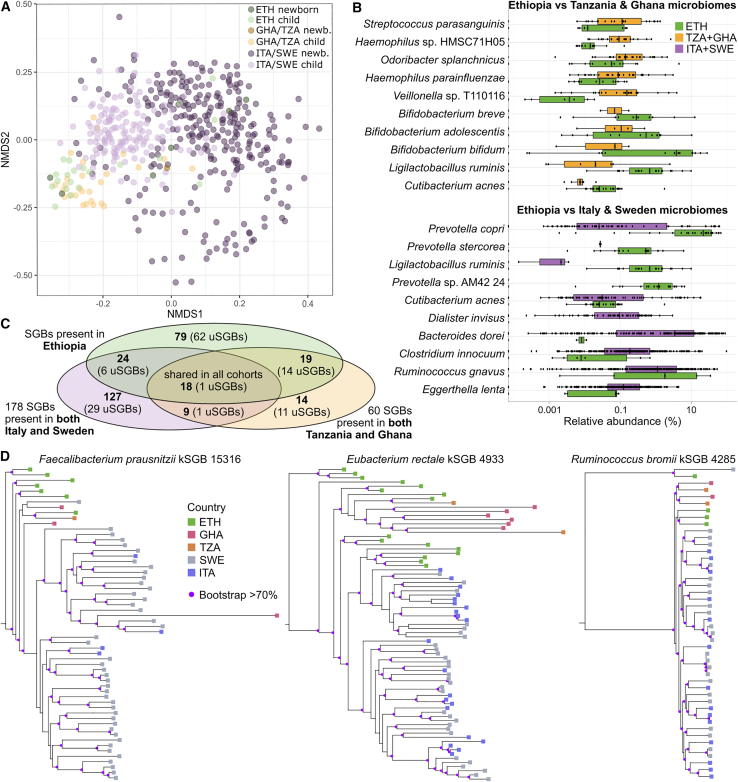
Figure 1.
Ethiopian mother and infant microbiome composition differs from that of age-matched Italian and Swedish subjects but resembles that of the two Tanzanian and Ghanaian populations
(A) Non-metric multidimensional scaling of the Bray-Curtis distance on sample taxonomic composition shows a trend ranging from newborns (<1 year of age, darker colors) to children (≥1 year of age, lighter colors). Ethiopian samples (green shades) tend to cluster together and with samples from the Tanzanian and Ghanaian communities (yellow shades), with the exception of some infant and child samples that cluster with same-age samples of the Italian and Swedish cohorts (violet shades). NMDS presenting the different cohorts, including adult samples, is available in Figure S1A.
(B) Species most associated with Ethiopian, westernized (here, Italian and Swedish), and non-westernized (here, Tanzanian and Ghanaian) infants aged 0–12 years. Because of the different age distribution between non-westernized and westernized infants, we randomly picked westernized infants aged 0–6 months, 6–12 months, and 1–12 years to get a similar age distribution with respect to Ethiopian ones before performing a Wilcoxon test. The test was repeated 10 times with 10 different random pickings, with each test comparing the microbiome composition of 127 westernized and 20 Ethiopian infants (0–6 months: 55 West. and 12 Eth.; 6–12 months: 27 West. and 6 Eth.; 12–36 months: 9 West. and 2 Eth.). Non-westernized cohorts already had an age distribution similar to the Ethiopian one, so we performed a simple Wilcoxon test (28 non-West. and 26 Eth.). Reported here are the 20 species that showed the strongest association with Ethiopian, westernized, or non-westernized infants and that had >0.05% average relative abundance and >20% prevalence in at least one of the two compared categories. For a full list, see Data S1. The box plots show the first and third quartiles (boxes) and the median (middle line); the whiskers extend up to 1.5× the IQR.
(C) Ethiopian infants share more species-level genome bins (SGBs, spanning < 5% genetic diversity, see STAR Methods) and particularly more uSGBs (SGBs assigned to uncharacterized species) with infants from Tanzania and Ghana (22.7% of SGBs found in Ethiopian and in both non-westernized infants’ cohorts) and have only limited SGB overlap with Italian and Swedish infants’ microbiomes (15.2% of SGBs found in Ethiopian and in both westernized infants’ cohorts). Of these overlapping SGBs, only 18 are found at least once in infants from all countries, with only 1 uSGB shared across all cohorts (Eubacterium SGB4290; more details in Data S2C).
(D) Phylogenetic trees based on SGB-specific core genes (see STAR Methods) of the most relevant species shared across all infant cohorts included in this study (≥2 positive infants per cohort, SGBs taxonomically assigned to the same species are summed. Subtrees with bootstrap > 70% are identified with purple circles. The term kSGB indicates an SGB assigned to species for which reference genomes are available; uSGB indicates an SGB assigned to a species lacking reference genomes). Additional phylogenetic trees of interesting species are reported in Figure S1C.