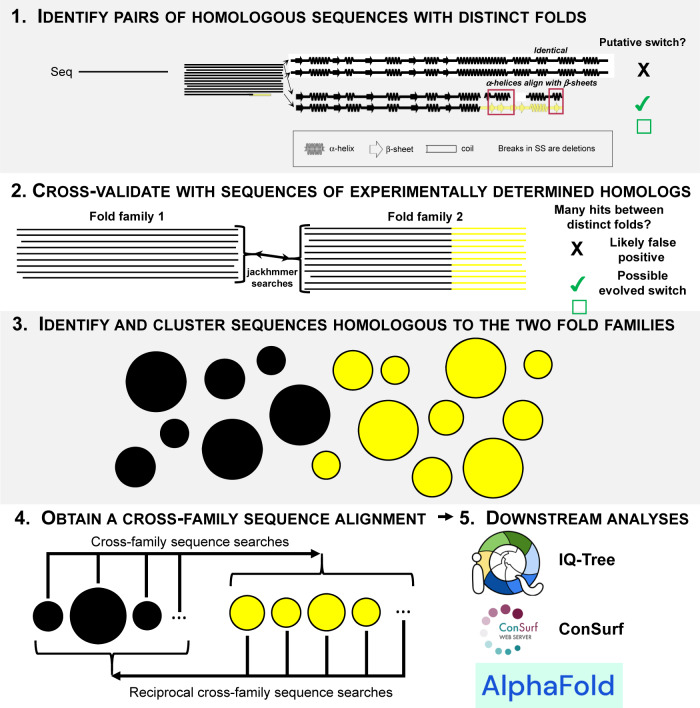
Fig. 6. A step-by-step guide to identifying more evolved fold switchers.
1. Query a sequence of interest (black) against the PDB (or database of predicted structures) with one round of protein BLAST (or phmmer) and search for hits with distinct secondary structures (yellow). Hits may indicate evolved fold switching. 2. Cross-validate results from step 1 by performing more sensitive sequence searches (e.g., jackhmmer) of all homologous sequences with experimentally determined structures. Black sequences=Fold1; yellow sequences=Fold2. Black regions of Fold2 have the same folds as Fold1 to allow for the possibility that Fold2 is a protein subdomain. 3. If cross validation is successful, find all sequences homologous to Fold1 (black) and Fold2 (yellow); cluster sequences by likely fold family. 4. Obtain a cross-family sequence alignment by searching all sequences from Fold1 against Fold2 and reciprocally searching Fold2 hits against Fold1. 5. Use cross-family alignment for downstream analyses including, but not limited to, IQ-Tree, ConSurf, and AlphaFold2. Complete descriptions of each step can be found in the main text.