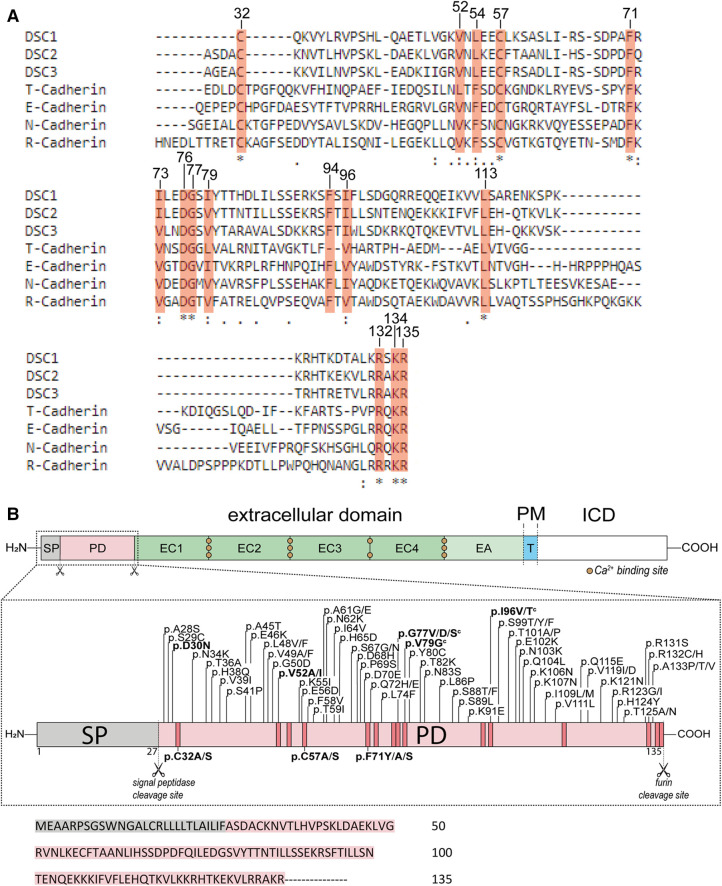
Figure 1.
(A) comparison of the prodomains (PD) of DSC1-3 and different classical cadherins (N-, R-, E-Cadherin) and cadherin-like proteins (T-Cadherin) from Homo sapiens using Clustal Omega (28). Conserved amino acids are marked with orange boxes. Positions on top refer to the open reading frame (ORF) of DSC2. (B) Top: Schematic overview of the structure of DSC2. The extracellular domain consists of the signal peptide (SP, grey box), the PD (light orange box), four extracellular cadherin domains (EC1-4, green boxes) and an anchor domain (EA, light green box) which is sometimes assigned as a fifth cadherin domain (EC5). A single transmembrane domain (T, blue box) connects the extracellular region with the intracellular domain (ICD, white box). PM, plasma membrane. Middle: Schematic overview of the SP and PD of DSC2. Missense variants that are associated with ACM are shown on top and were received from the ARVC database (29), the Human Gene Mutation Database (HGMD) (30), from the National Library of Medicine ClinVar (31) and the genome aggregation database (gnomAD) (32). Variants below are investigated model mutants. All variants are classified as variants with unknown significance (VUS) according to the ACMG guidelines (27). Variants shown in bold letters were investigated. Scissors represent cleavage sites. Orange boxes mark highly conserved amino acids. c indicates variants that share conflicting interpretation of pathogenicity in various genetic databases. Bottom: Primary amino acid sequence of the SP and PD of human DSC2.