Figure 2.
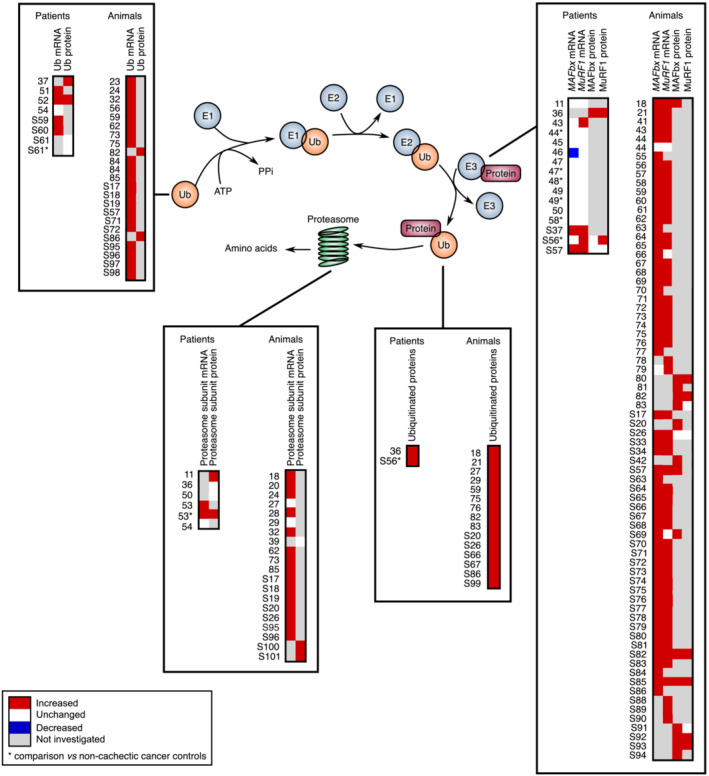
Comparative analysis of the transcript and protein levels of components of the ubiquitin–proteasome system in cachectic skeletal muscle of cancer patients and animals. Proteins are targeted for degradation by the 26S proteasome through covalent attachment of a chain of ubiquitin molecules. The E1 ubiquitin‐activating enzyme hydrolyses ATP to bind ubiquitin. E2 ubiquitin‐conjugating enzymes receive ubiquitin from E1 and brings it to the E3 ubiquitin‐ligase enzymes, which catalyse the transfer of the ubiquitin from E2 to the substrate. This reaction is the rate‐limiting step of the ubiquitination process. The ubiquitinated protein is then docked to the proteasome for degradation. One gene encodes the E1 enzyme, whereas one hundred genes encode the E2 enzymes and almost one thousand genes the E3 enzymes. Significant variations are reported in red (increase) or blue (decrease). Unchanged levels are reported in white.
